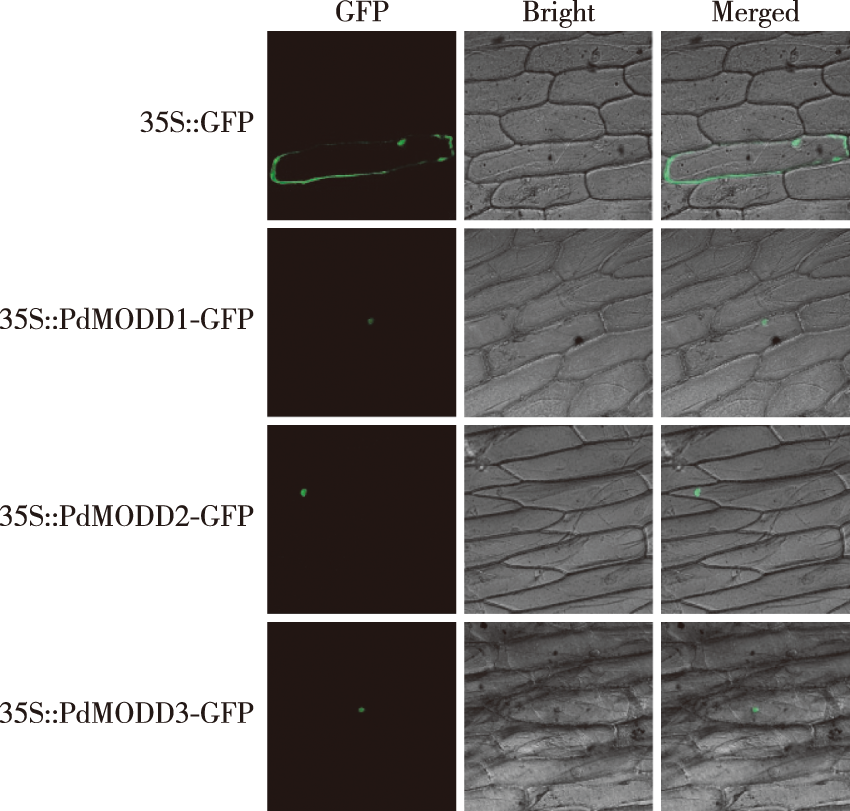
【目的】研究杨树重要胁迫响应新基因,为揭示杨树抗逆分子机制、培育优良抗逆杨树新品种提供理论依据。【方法】基于水稻MODD氨基酸序列,通过BLAST在美洲黑杨基因组中筛选得到3条基因(Podel.08G114100、Podel.10G158900和Podel.17G098500),并以其序列为参考,以‘渤丰3号’杨cDNA为模板克隆得到3条基因(PdMODD1、PdMODD2和PdMODD3)。利用生物信息学分析PdMODD蛋白的结构特征及与其同源蛋白的亲缘关系。通过基因枪轰击法分析PdMODD基因亚细胞定位。采用实时荧光定量(qRT-PCR)技术,分析PdMODD基因组织特异性和不同胁迫条件下的的表达模式。【结果】PdMODD1~3基因的开放阅读框(ORF)全长分别为1 065、1 065和1 074 bp,编码354、354和357个氨基酸,均为不稳定的亲水性蛋白。PdMODD基因均为NINJA家族成员,含有3个保守结构域,其中一个为转录抑制基序EAR。系统进化树分析显示,PdMODD1蛋白与毛果杨PtAFP2亲缘性较高并与拟南芥AtAFP1和木薯MeAFP2位于同一个分支;PdMODD2蛋白与麻疯树JcAFP2亲缘关系最近,并与拟南芥AtAFP2聚成一个分支;PdMODD3与毛果杨PtAFP3-1亲缘关系最近,与栓皮槠QsAFP3、拟南芥AtAFP4属于同一分支。亚细胞定位结果表明PdMODD基因均定位于细胞核。组织特异性分析显示,PdMODD1~3在根、茎、叶中均能表达,且在叶中表达量最高,根中表达量最低。胁迫响应表达模式结果显示,NaCl和聚乙二醇(PEG)胁迫处理下,PdMODD1~3在根、茎、叶组织中主要为显著上调表达。【结论】PdMODD1~3均为NINJA家族成员,含有保守的转录抑制基序EAR,具有组织特异性,在叶中高表达,且能被NaCl和PEG胁迫显著诱导表达,推测PdMODD1、PdMODD2 和PdMODD3可能会在‘渤丰3号’杨对盐和干旱胁迫响应中发挥重要的调节作用。
【Objective】To explore the function of PdMODD in the growth and development of Populus × euramericana, we analyzed their sequence characteristics, subcellular localization and expression patterns in response to salt and drought stress, which will provide a theoretical basis for revealing the molecular mechanisms of stress resistance and breeding new poplar varieties. 【Method】Based on the rice MODD amino acid sequence, Podel.08G114100, Podel.10G158900 and Podel.17G098500 were selected using BLAST in the genome of Populus deltoides WV94 v2.1. Using these gene sequences as references and Populus × euramericana ‘Bofeng 3 hao’ cDNA as template, three genes (PdMODD1,PdMODD2 and PdMODD3) were cloned. The structural characteristics of the PdMODD protein and its genetic relationship with its homologous proteins were analyzed using bioinformatics methods. The subcellular localization of PdMODD genes was analyzed using particle bombardment. The expression characteristics of PdMODD genes in different tissues and their response to different stresses were analyzed using qRT-PCR. 【Results】The open reading frames of PDMODD1-PDMODD3 genes were 1 065, 1 065 and 1 074 bp, encoding 354, 354 and 357 amino acids, respectively, all of which were unstable hydrophilic proteins located in the nucleus.PdMODD genes are all members of the NINJA family and contain three conserved domains, one of which is the transcriptional repression motif EAR. Phylogenetic analysis demonstrated that PdMODD1 protein was closely related to PtAFP2 (Populus trichocarpa) and located in the same branch as AtAFP1 (Arabidopsis thaliana) and MeAFP2 (Manihot esculenta). PdMODD2 protein had a close relationship with JcAFP2 (Jatropha curcas) and converged into a branch with AtAFP2; PdMODD3 was closely related to PtAFP3-1 and located in the same branch as QsAFP3 (Quercus suber) and AtAFP4. The subcellular localization results showed that PdMODD genes were localized in the nucleus. The results of tissue specificity analysis showed that PdMODD genes were highly expressed in the leaves and the least expressed in the roots. The expression patterns of stress response showed that under NaCl and PEG stress, PdMODD1, PdMODD2 and PdMODD3 were significantly upregulated in the root, stem and leaf tissues. 【Conclusion】PdMODD1-PdMODD3 are all members of the NINJA family, with the conserved transcriptional repression motif EAR and tissue specificity. They were highly expressed in the leaves, significantly induced by NaCl and PEG stress, and were in a branch with AtAFP1, AtAFP2 and AtAFP4, which had negative regulatory effects on ABA and salt stress. It is hypothesized that PdMODD1, PdMODD2 and PdMODD3 may play an important regulatory role in response to P. euramericana ‘Bofeng 3 hao’ salt and drought stress.
 PDF(4500 KB)
PDF(4500 KB)


 PDF(4500 KB)
PDF(4500 KB)
 PDF(4500 KB)
PDF(4500 KB)