 PDF(3438 KB)
PDF(3438 KB)


植物功能性状系统发育保守性的类群和地理分异研究——以中国被子植物最大株高为例
邢冰冰, 李垚, 毛岭峰
南京林业大学学报(自然科学版) ›› 2024, Vol. 48 ›› Issue (1) : 59-66.
 PDF(3438 KB)
PDF(3438 KB)
 PDF(3438 KB)
PDF(3438 KB)
植物功能性状系统发育保守性的类群和地理分异研究——以中国被子植物最大株高为例
Taxonomic and geographic differentiation of phylogenetic conservatism of plant functional traits: a case study of maximum plant height of Chinese angiosperms
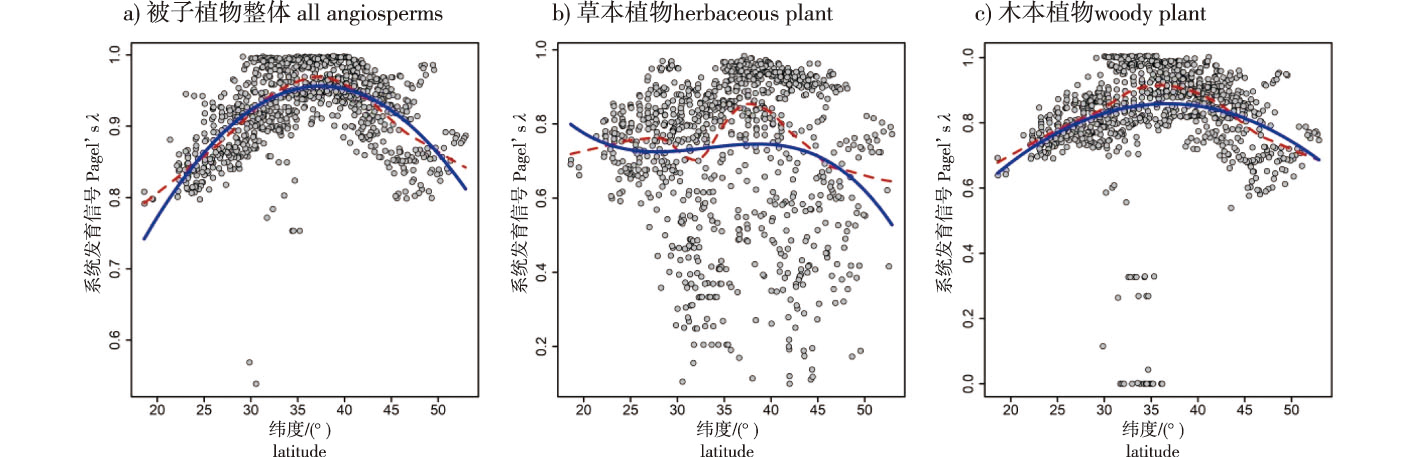
【目的】株高是植物生态策略的核心部分。本研究旨在揭示中国被子植物最大株高的系统发育保守性,探明其类群、地理分异规律及与环境因子的关联。【方法】利用中国20 295种被子植物的最大株高和地理分布数据,检测不同生长型、不同类群和不同植被区域植物最大株高的系统发育保守性,分析系统发育信号(Pagel’s λ值)与纬度、温度和降水量等因子的关联。【结果】中国被子植物最大株高的系统发育保守性较强(Pagel’s λ = 0.893)。其中,草本植物的系统发育保守性程度略低于木本植物,豆目(Fabales)、石竹目(Caryophyllales)、天门冬目(Asparagales)、唇形目(Lamiales)、伞形目(Apiales)、虎耳草目(Saxifragales)等6个目的系统发育信号均高于0.9。所有被子植物与木本植物的系统发育信号与纬度、温度均呈单峰曲线关系,但与降水量呈负相关。草本植物系统发育信号的纬度格局不明显,与温度、降水量均呈多峰曲线关系。【结论】中国被子植物最大株高的系统发育保守性具有明显的地理格局,但趋势因生长型而异。总体来说,中纬度地区被子植物整体及木本植物株高的系统发育保守性强于高、低纬度地区。
【Objective】 Plant height is a fundamental component of plant ecological strategies. This study aims to elucidate the phylogenetic conservatism of maximum plant height in Chinese angiosperms, investigate its variation across taxonomic groups and geographic regions, and explore its associations with environmental factors. 【Method】 Using data on the maximum plant height and geographic distribution of 20 295 Chinese angiosperm species, this study assessed the phylogenetic conservatism of maximum plant height among different growth forms, taxonomic groups, and vegetation zones. The study analyzed the correlation between the phylogenetic signal (Pagel’s λ) and factors such as latitude, temperature, and precipitation. 【Result】 The maximum plant height of Chinese angiosperms exhibited a strong phylogenetic conservatism (Pagel’s λ = 0.893). Among them, herbaceous plants showed slightly lower phylogenetic conservatism compared to woody plants, while six orders including Fabales exhibited phylogenetic signals higher than 0.9. The phylogenetic signals of all plants and woody plants displayed a unimodal relationship with latitude and temperature but showed a negative correlation with precipitation. The latitudinal pattern of phylogenetic signals in herbaceous plants was less pronounced, instead showing a multi-peak relationship with temperature and precipitation. 【Conclusion】 The maximum plant height of Chinese angiosperms demonstrated a clear geographic pattern in terms of phylogenetic conservatism, but the trend varied among different growth forms. Overall, the phylogenetic conservatism of maximum plant height of angiosperms and woody plants was stronger in middle latitudes compared to both high and low latitudes.
被子植物 / 功能性状 / 系统发育信号 / 系统发育保守性 / 最大株高 / 中国
angiosperm / functional trait / phylogenetic signal / phylogenetic conservatism / maximum plant height / China
| [1] |
|
| [2] |
霍佳璇, 任梁, 潘莹萍, 等. 柴达木盆地荒漠植物功能性状及其对环境因子的响应[J]. 生态学报, 2022, 42(11):4494-4503.
|
| [3] |
贾婷, 宋武云, 关新贤, 等. 湿地松针叶功能性状及其对磷添加的响应[J]. 南京林业大学学报(自然科学版), 2021, 45(6):65-71.
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
许格希, 史作民, 刘顺, 等. 尖峰岭热带山地雨林林冠层乔木某些功能性状的系统发育信号、关联性及其演化模式[J]. 生态学报, 2017, 37(17):5691-5703.
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
邱思玉, 曹元帅, 孙玉军, 等. 杉木人工林与年龄无关的优势高生长模型[J]. 南京林业大学学报(自然科学版), 2019, 43(5): 121-127.
|
| [35] |
毛岭峰. 中国种子植物多样性的空间格局-环境关系分异研究[D]. 北京: 中国科学院植物研究所, 2013.
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
TAP Group. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV[J]. Bot J Linn Soc, 2016, 181(1):1-20. DOI: 10.1111/boj.12385.
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
郄亚栋, 蒋腊梅, 吕光辉, 等. 温带荒漠植物叶片功能性状对土壤水盐的响应[J]. 生态环境学报, 2018, 27(11):2000-2010.
|
| [49] |
|
| [50] |
邵晨, 李耀琪, 罗奥, 等. 不同生活型被子植物功能性状与基因组大小的关系[J]. 生物多样性, 2021, 29(5):575-585.
|
| [51] |
袁泉, 曹嘉瑜, 刘建峰, 等. 生长型分类方案不同导致森林生态系统植物功能性状的统计偏差[J]. 生态学报, 2021, 41(3):1106-1115.
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
/
| 〈 |
|
〉 |