 PDF(6238 KB)
PDF(6238 KB)


 PDF(6238 KB)
PDF(6238 KB)
 PDF(6238 KB)
PDF(6238 KB)
小果白刺bHLH基因家族鉴定与表达分析
Identification and expression analysis of the bHLH gene family in Nitraria sibirica
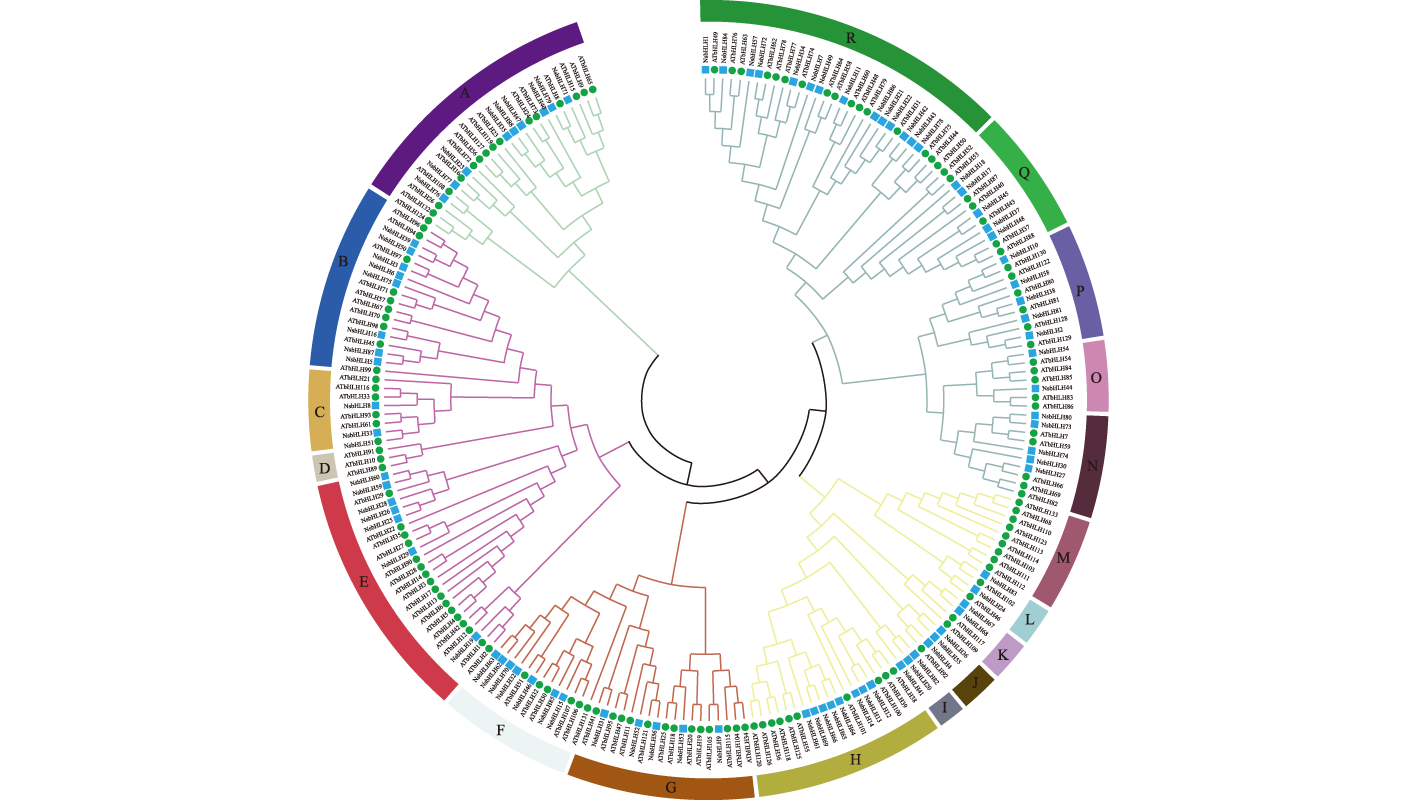
【目的】基本螺旋-环-螺旋(bHLH)转录因子在植物的生长、代谢和对生物/非生物胁迫的响应等过程中发挥着重要作用。进行小果白刺(Nitraria sibirica)bHLH全基因组鉴定与生物信息学分析,可为后续bHLH基因克隆和进一步生物学功能研究提供参考。【方法】用生物信息学和分子生物学方法,在全基因组水平对小果白刺bHLH基因家族进行系统进化分析并利用转录组数据及定量qRT-PCR分析小果白刺bHLH基因家族对盐胁迫的响应。【结果】基于小果白刺基因组数据共鉴定出88个bHLH转录因子,不均匀地分布在12条染色体上,并有2对串联重复基因,根据系统进化树可将其划分为16亚群;顺式作用元件分析显示,88个bHLH转录因子含有多种与环境和胁迫相关的顺式作用元件;基因表达模式分析表明,小果白刺bHLHs在盐胁迫过程中响应程度不一,qRT-PCR结果显示6个bHLH基因在不同组织中表达特异性显著;盐胁迫下,bHLH1、bHLH24、bHLH70、bHLH73在叶中表达显著上调,而bHLH2、bHLH30在叶中的表达下调;在根中这6个基因表现出相同的表达模式,均显著上调;在茎中除了bHLH2有下调表达外,其他5个基因均为上调表达。【结论】鉴定出88个小果白刺bHLH转录因子,其基因结构和结构域较保守,染色体上有2对串联重复基因;盐胁迫处理后,小果白刺bHLH基因家族成员发生响应,在根、茎和叶中均有表达,可能与该家族成员具有抗盐功能有关。该研究为进一步阐明小果白刺bHLH转录因子家族基因的功能奠定基础。
【Objective】The basic helix-loop-helix (bHLH) transcription factors play critical roles in plant growth, metabolism, and responses to biotic/abiotic stresses. Genome-wide identification and bioinformatics analysis of the bHLH gene family in Nitraria sibirica will provide foundational insights for subsequent bHLH gene cloning and further biological studies.【Method】Bioinformatics and molecular biology approaches were employed to systematically analyze the phylogenetic relationships of the N. sibirica bHLH gene family at the genome-wide level. Transcriptome datasets and quantitative reverse transcription-PCR (qRT-PCR) were employed to analyze the response of the bHLH genes under to stress.【Result】A total of 88 bHLH transcription factors were identified from the N. sibirica genome, unevenly distributed across 12 chromosomes with two pairs of tandemly duplicated gene. Phylogenetic analysis classified these members into 16 distinct subclades. Cis-acting element analysis revealed that the 88 bHLH genes harbored multiple environment- and stress-responsive regulatory elements. Expression profiling demonstrated differential responses of N. sibirica bHLH genes to salt stress. qRT-PCR results further demonstrated that six bHLH genes (bHLH1, bHLH2, bHLH24, bHLH30, bHLH70 and bHLH73) exhibited pronounced tissue-specific expression patterns. Under salt stress, bHLH1, bHLH24, bHLH70 and bHLH73 were significantly upregulated in leaves, while bHLH2 and bHLH30 were down regulated in leaves. In roots, all six genes displayed marked upregulation. In stems, five genes (bHLH1, bHLH24, bHLH30, bHLH70 and bHLH73) were upregulated, whereas bHLH2 showed down regulation.【Conclusion】This study identified 88 conserved bHLH transcription factors in N. sibirica, characterized by conserved gene structures and protein domains, along with two tandem duplication events on chromosomes. The differential expression of bHLH genes in roots, stems and leaves under salt stress suggests their potential involvement in salt tolerance mechanisms. These findings establish a critical foundation for elucidating the functional roles of the bHLH transcription factor family in N. sibirica.
Nitraria sibirica / bHLH transcription factors / salt stress / gene expression
| [1] |
朱涛, 李芳菲, 杨海涵, 等. 山药bHLH基因家族鉴定及表达分析[J]. 信阳师范学院学报(自然科学版), 2022, 35(3):393-399.
|
| [2] |
The basic Helix-Loop-Helix (bHLH) proteins are transcription factors that play important roles during the development of various metazoans including fly, nematode, and vertebrates. They are also involved in human diseases, particularly in cancerogenesis. We made an extensive search for bHLH sequences in the completely sequenced genomes of Caenorhabditis elegans and of Drosophila melanogaster. We found 35 and 56 different genes, respectively, which may represent the complete set of bHLH of these organisms. A phylogenetic analysis of these genes, together with a large number (>350) of bHLH from other sources, led us to define 44 orthologous families among which 36 include bHLH from animals only, and two have representatives in both yeasts and animals. In addition, we identified two bHLH motifs present only in yeast, and four that are present only in plants; however, the latter number is certainly an underestimate. Most animal families (35/38) comprise fly, nematode, and vertebrate genes, suggesting that their common ancestor, which lived in pre-Cambrian times (600 million years ago) already owned as many as 35 different bHLH genes.
|
| [3] |
|
| [4] |
A natural (evolutionary) classification is provided for 242 basic helix-loop-helix (bHLH) motif-containing proteins. Phylogenetic analyses of amino acid sequences describe the patterns of evolutionary change within the motif and delimit evolutionary lineages. These evolutionary lineages represent well known functional groups of proteins and can be further arranged into five groups based on binding to DNA at the hexanucleotide E-box, the amino acid patterns in other components of the motif, and the presence/absence of a leucine zipper. The hypothesized ancestral amino acid sequence for the bHLH transcription factor family is given together with the ancestral sequences of the subgroups. It is suggested that bHLH proteins containing a leucine zipper are not a natural, monophyletic group.
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
Tobacco (Nicotiana tabacum) is a member of the Solanaceae, one of the agronomically most important groups of flowering plants. We have performed an in silico analysis of 1.15 million gene-space sequence reads from the tobacco nuclear genome and report the detailed analysis of more than 2,500 tobacco transcription factors (TFs). The tobacco genome contains at least one member of each of the 64 well-characterized TF families identified in sequenced vascular plant genomes, indicating that evolution of the Solanaceae was not associated with the gain or loss of TF families. However, we found notable differences between tobacco and non-Solanaceae species in TF family size and evidence for both tobacco- and Solanaceae-specific subfamily expansions. Compared with TF families from sequenced plant genomes, tobacco has a higher proportion of ERF/AP2, C2H2 zinc finger, homeodomain, GRF, TCP, zinc finger homeodomain, BES, and STERILE APETALA (SAP) genes and novel subfamilies of BES, C2H2 zinc finger, SAP, and NAC genes. The novel NAC subfamily, termed TNACS, appears restricted to the Solanaceae, as they are absent from currently sequenced plant genomes but present in tomato (Solanum lycopersicum), pepper (Capsicum annuum), and potato (Solanum tuberosum). They constitute approximately 25% of NAC genes in tobacco. Based on our phylogenetic studies, we predict that many of the more than 50 tobacco group IX ERF genes are involved in jasmonate responses. Consistent with this, over two-thirds of group IX ERF genes tested showed increased mRNA levels following jasmonate treatment. Our data are a major resource for the Solanaceae and fill a void in studies of TF families across the plant kingdom.
|
| [9] |
The first decision made by an angiosperm seed, whether to germinate or not, is based on integration of various environmental signals such as water and light. The phytochromes (Phys) act as red and far-red light (Pfr) photoreceptors to mediate light signaling through yet uncharacterized pathways. We report here that the PIF3-like 5 (PIL5) protein, a basic helix-loop-helix transcription factor, is a key negative regulator of phytochrome-mediated seed germination. PIL5 preferentially interacts with the Pfr forms of Phytochrome A (PhyA) and Phytochrome B (PhyB). Analyses of a pil5 mutant in conjunction with phyA and phyB mutants, a pif3 pil5 double mutant, and PIL5 overexpression lines indicate that PIL5 is a negative factor in Phy-mediated promotion of seed germination, inhibition of hypocotyl negative gravitropism, and inhibition of hypocotyl elongation. Our data identify PIL5 as the first Phy-interacting protein that regulates seed germination.
|
| [10] |
|
| [11] |
Several processes of plant development, such as abscission, pollen release, fruit dehiscence, and seed dispersal, require organs or tissues to physically disassociate or split open. Due to the immobility of plant cells, these processes occur through coordinated mechanisms of cell separation that are not found in animals. Arabidopsis produces dry dehiscent fruits (siliques) making it a convenient system for the genetic study of cell separation associated with dehiscence.We describe here a novel mutation in Arabidopsis called alcatraz (alc), which prevents dehiscence of fruit by specifically blocking the separation of the valve cells from the replum. The ALC gene is shown to encode a protein related to the myc/bHLH family of transcription factors and is expressed in the valve margins of the silique, which is the site of cell separation during dehiscence. Detailed studies using TEM indicates that ALC enables cell separation in Arabidopsis fruit dehiscence by promoting the differentiation of a strip of labile nonlignified cells sandwiched between layers of lignified cells. Transgenic plants expressing antisense or dominant-negative ALC are defective in silique dehiscence.Cell separation in fruit dehiscence requires a specialized cell layer which is nonlignified and capable of autolysis, specified by a myc/bHLH protein encoded by ALC. These findings may have relevance to other processes requiring cell separation, as well as for the practical design of crops with reduced seed losses.
|
| [12] |
|
| [13] |
任镘蓉, 全英杰, 杨雯婷, 等. 地被菊bHLH转录因子基因家族鉴定及其低温胁迫响应[J]. 农业生物技术学报, 2022, 30(1):50-62.
|
| [14] |
|
| [15] |
Ethylene signaling pathways regulate several physiological alterations that occur during tomato fruit ripening, such as changes in colour and flavour. The mechanisms underlying the transcriptional regulation of genes in these pathways remain unclear, although the role of the MADS-box transcription factor RIN has been widely reported. Here, we describe a bHLH transcription factor, SlbHLH95, whose transcripts accumulated abundantly in breaker+4 and breaker+7 fruits compared with rin (ripening inhibitor) and Nr (never ripe) mutants. Moreover, the promoter activity of SlbHLH95 was regulated by RIN in vivo. Suppression of SlbHLH95 resulted in reduced sensitivity to ethylene, decreased accumulation of total carotenoids, and lowered glutathione content, and inhibited the expression of fruit ripening- and glutathione metabolism-related genes. Conversely, up-regulation of SlbHLH95 in wild-type tomato resulted in higher sensitivity to ethylene, increased accumulation of total carotenoids, slightly premature ripening, and elevated accumulation of glutathione, soluble sugar, and starch. Notably, overexpression of SlbHLH95 in rin led to the up-regulated expression of fruit ripening-related genes (FUL1, FUL2, SAUR69, ERF4, and CNR) and multiple glutathione metabolism-related genes (GSH1, GSH2, GSTF1, and GSTF5). These results clarified that SlbHLH95 participates in the regulation of fruit ripening and affects ethylene sensitivity and multiple metabolisms targeted by RIN in tomato.© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Experimental Biology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
|
| [16] |
|
| [17] |
何洁, 顾秀容, 魏春华, 等. 西瓜bHLH转录因子家族基因的鉴定及其在非生物胁迫下的表达分析[J]. 园艺学报, 2016, 43(2):281-294.
利用生物信息学方法,在西瓜测序基因组97103 中共鉴定出96 个bHLH 家族成员,其中有94 个可以被定位到西瓜的11 条染色体上。通过基因结构和结构域序列保守性的预测,发现这些基因的序列长度和内含子数量变化很大,但bHLH 结构域序列比较保守。用拟南芥中39 条已知的bHLH 蛋白序列和西瓜的96 条bHLH 蛋白序列构建系统发育树,结果显示西瓜的bHLH 家族可以进一步被分为11 个亚族。运用荧光定量实时PCR 技术,分析了该家族中21 个基因在西瓜响应非生物胁迫时的表达水平,结果表明,8 个基因受低温胁迫诱导表达,13 个基因受ABA胁迫诱导表达,14 个基因受盐胁迫诱导表达。ClabHLH41在3 种胁迫下表达量均显著增加,说明其在低温、ABA 和盐胁迫应答中可能发挥着重要作用。
|
| [18] |
耿晶晶. 甜橙bHLH家族转录因子发掘及CsbHLH18抗寒功能鉴定与作用机制解析[D]. 武汉: 华中农业大学, 2018.
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
李焕勇, 杨秀艳, 唐晓倩, 等. NaCl处理对小果白刺叶片主要渗透调节物质和激素水平的影响[J]. 东北林业大学学报, 2019, 47(5):30-35.
|
| [23] |
胡娟, 张洁, 盛洲, 等. 不同预处理对柴达木盆地高寒湿地小果白刺种子萌发的影响[J]. 种子, 2022, 41(3):87-92,2.
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
Plant basic/helix–loop–helix (bHLH) transcription factors participate in a number of biological processes, such as growth, development and abiotic stress responses. The bHLH family has been identified in many plants, and several bHLH transcription factors have been functionally characterized in Arabidopsis. However, no systematic identification of bHLH family members has been reported in potato (Solanum tuberosum). Here, 124 StbHLH genes were identified and named according to their chromosomal locations. The intron numbers varied from zero to seven. Most StbHLH proteins had the highly conserved intron phase 0, which accounted for 86.2% of the introns. According to the Neighbor-joining phylogenetic tree, 259 bHLH proteins acquired from Arabidopsis and potato were divided into 15 groups. All of the StbHLH genes were randomly distributed on 12 chromosomes, and 20 tandem duplicated genes and four pairs of duplicated gene segments were detected in the StbHLH family. The gene ontology (GO) analysis revealed that StbHLH mainly function in protein and DNA binding. Through the RNA-seq and quantitative real time PCR (qRT-PCR) analyses, StbHLH were found to be expressed in various tissues and to respond to abiotic stresses, including salt, drought and heat. StbHLH1, 41 and 60 were highly expressed in flower tissues, and were predicted to be involved in flower development by GO annotation. StbHLH45 was highly expressed in salt, drought and heat stress, which suggested its important role in abiotic stress response. The results provide comprehensive information for further analyses of the molecular functions of the StbHLH gene family.
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
尚先文, 范付华, 周紫晶, 等. 马尾松苗期转录组bHLH基因家族成员鉴定及表达分析[J]. 农业生物技术学报, 2020, 28(11):1947-1959.
|
| [33] |
赵安琪, 张世杰, 张志国, 等. 海水胁迫下萱草bHLH转录因子家族的鉴定及表达分析[J]. 分子植物育种, 2025, 23(16):5332-5343.
|
| [34] |
陈微, 潘美红, 惠林冲, 等. 基于转录组的洋葱bHLH转录因子家族鉴定与生物信息学分析[J]. 江苏农业科学, 2023, 51(5):11-18.
|
| [35] |
廖桂花, 段钰, 王丛丛, 等. 赪桐bHLH转录因子家族鉴定及生物信息学分析[J/OL]. 分子植物育种, 2023: 1-17. (2025-03-15).
|
| [36] |
\n The basic helix-loop-helix (bHLH) gene family is one of the largest transcription factor families in plants and is functionally characterized in diverse species. However, less is known about its functions in the economically important allopolyploid oil crop,\n Brassica napus\n.\n
|
| [37] |
|
| [38] |
BackgroundIn plants, the basic helix-loop-helix (bHLH) transcription factors play key roles in diverse biological processes. Genome-wide comprehensive and systematic analyses of bHLH proteins have been well conducted in Arabidopsis, rice, tomato and other plant species. However, only few of bHLH family genes have been functional characterized in maize.ResultsIn this study, our genome-wide analysis identified 208 putative bHLH family proteins (ZmbHLH proteins) in maize (Zea mays). We classified these proteins into 18 subfamilies by comparing the ZmbHLHs with Arabidopsis thaliana bHLH proteins. Phylogenetic analysis, conserved protein motifs, and exon-intron patterns further supported the evolutionary relationships among these bHLH proteins. Genome distribution analysis found that the 208 ZmbHLH loci were located non-randomly on the ten maize chromosomes. Further, analysis of conserved cis-elements in the promoter regions, protein interaction networks, and expression patterns in roots, leaves, and seeds across developmental stages, suggested that bHLH family proteins in maize are probably involved in multiple physiological processes in plant growth and development.ConclusionWe performed a genome-wide, systematic analysis of bHLH proteins in maize. This comprehensive analysis provides a useful resource that enables further investigation of the physiological roles and molecular functions of the ZmbHLH transcription factors.
|
| [39] |
冯建英, 李立芹, 鲁黎明. 马铃薯bHLH转录因子家族全基因组鉴定与表达分析[J]. 生物技术通报, 2022, 38(2):21-33.
对马铃薯bHLH转录因子进行全基因组鉴定与表达模式分析,为其生物学功能研究提供借鉴。基于马铃薯基因组数据库和Pfam数据库,运用HMMER对马铃薯bHLH家族成员进行全基因组鉴定,剔除冗余序列后,利用ExPASy和MEME软件对候选序列进行基本理化性质及保守元件分析,并使用MEGA-X软件进行聚类分析;用MG2C和TBtools软件分别绘制染色体定位图和表达模式图。结果表明,从马铃薯基因组数据库中共检索出108个bHLH转录因子,其氨基酸数目为62-694个,分子量为7 527.78-75 939.94 Da,理论等电点为4.55-10.40。这些bHLH转录因子分布在马铃薯的12条染色体上,均具有典型的HLH结构域,可划分为16个亚族。基因表达模式分析表明,马铃薯bHLH家族成员具有组织表达特异性,多数响应盐、甘露醇、生长素、脱落酸、赤霉素及热等胁迫。马铃薯全基因组包括108个高度保守的bHLH基因家族成员,其表达具有组织特异性,并且响应非生物胁迫的诱导。
|
| [40] |
张尚宏, 屈良鹄. 基因组的进化与内含子中的基因的进化[J]. 中山大学学报(自然科学版), 1999, 38(1):49-53.
|
| [41] |
Gene duplication plays key roles in organismal evolution. Duplicate genes, if they survive, tend to diverge in regulatory and coding regions. Divergences in coding regions, especially those that can change the function of the gene, can be caused by amino acid-altering substitutions and/or alterations in exon-intron structure. Much has been learned about the mode, tempo, and consequences of nucleotide substitutions, yet relatively little is known about structural divergences. In this study, by analyzing 612 pairs of sibling paralogs from seven representative gene families and 300 pairs of one-to-one orthologs from different species, we investigated the occurrence and relative importance of structural divergences during the evolution of duplicate and nonduplicate genes. We found that structural divergences have been very prevalent in duplicate genes and, in many cases, have led to the generation of functionally distinct paralogs. Comparisons of the genomic sequences of these genes further indicated that the differences in exon-intron structure were actually accomplished by three main types of mechanisms (exon/intron gain/loss, exonization/pseudoexonization, and insertion/deletion), each of which contributed differently to structural divergence. Like nucleotide substitutions, insertion/deletion and exonization/pseudoexonization occurred more or less randomly, with the number of observable mutational events per gene pair being largely proportional to evolutionary time. Notably, however, compared with paralogs with similar evolutionary times, orthologs have accumulated significantly fewer structural changes, whereas the amounts of amino acid replacements accumulated did not show clear differences. This finding suggests that structural divergences have played a more important role during the evolution of duplicate than nonduplicate genes.
|
| [42] |
Basic helix-loop-helix (bHLH) proteins are a class of transcription factors found throughout eukaryotic organisms. Classification of the complete sets of bHLH proteins in the sequenced genomes of Arabidopsis thaliana and Oryza sativa (rice) has defined the diversity of these proteins among flowering plants. However, the evolutionary relationships of different plant bHLH groups and the diversity of bHLH proteins in more ancestral groups of plants are currently unknown. In this study, we use whole-genome sequences from nine species of land plants and algae to define the relationships between these proteins in plants. We show that few (less than 5) bHLH proteins are encoded in the genomes of chlorophytes and red algae. In contrast, many bHLH proteins (100-170) are encoded in the genomes of land plants (embryophytes). Phylogenetic analyses suggest that plant bHLH proteins are monophyletic and constitute 26 subfamilies. Twenty of these subfamilies existed in the common ancestors of extant mosses and vascular plants, whereas six further subfamilies evolved among the vascular plants. In addition to the conserved bHLH domains, most subfamilies are characterized by the presence of highly conserved short amino acid motifs. We conclude that much of the diversity of plant bHLH proteins was established in early land plants, over 440 million years ago.
|
| [43] |
高莉娟, 张正社, 文裕, 等. 象草全基因组bHLH转录因子家族鉴定及表达分析[J]. 草业学报, 2022, 31(3):47-59.
bHLH转录因子家族不仅参与了植物的生长发育,而且在植物响应逆境胁迫和次生代谢方面发挥着关键作用。在全基因组水平对重要饲草及能源植物象草的bHLH转录因子家族进行了鉴定及分析,并利用转录组数据及定量RT-PCR分析了象草bHLH转录因子对赤霉素(GA<sub>3</sub>)和多效唑(PAC)的响应。结果显示:在象草中共鉴定出229个具有完整保守结构域的bHLH基因家族成员(CpbHLH001~CpbHLH229),不均匀地分布于14条染色体上;系统进化分析结果表明,229个CpbHLHs可被分为18个亚类,其中C亚类的成员数量最多,为41个;此外,相同亚家族中的大多数基因具有相似的基因结构和保守基序;基于转录组数据的表达谱分析结果发现,多数bHLH基因在象草茎尖组织中均对GA<sub>3</sub>和PAC有响应。随机挑选9个表达量较高的基因进一步通过qPCR进行验证。结果显示,经外源GA<sub>3</sub>和PAC处理之后,这9个基因因不同处理而差异表达,表明这9个基因可能与GA<sub>3</sub>和PAC介导的信号通路有关。综上所述,本研究为象草bHLH转录因子家族的生物学功能奠定了基础。
|
| [44] |
|
| [45] |
|
| [46] |
|
/
| 〈 |
|
〉 |