 PDF(1847 KB)
PDF(1847 KB)


资源植物黑老虎的比较叶绿体基因组学研究
翟学昌, 彭丽, 颜海飞, 朱柯帆, 张淑燕, 张彩云, 鲁显楷
南京林业大学学报(自然科学版) ›› 2024, Vol. 48 ›› Issue (6) : 71-78.
 PDF(1847 KB)
PDF(1847 KB)
 PDF(1847 KB)
PDF(1847 KB)
资源植物黑老虎的比较叶绿体基因组学研究
Comparative chloroplast genomics of the important resource plant Kadsura coccinea
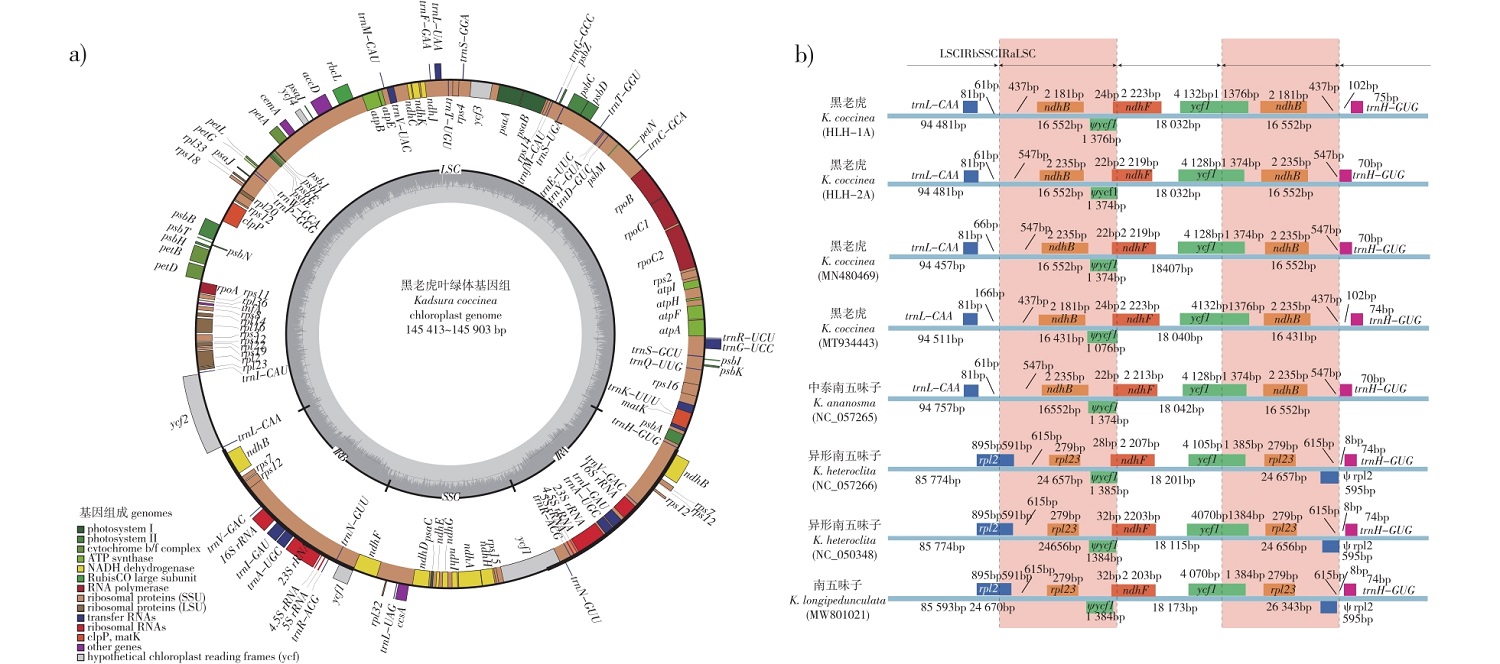
【目的】发掘我国重要资源植物黑老虎(Kadsura coccinea)的叶绿体基因组高变异区域及SSR变异位点,为其遗传多样性及种质资源评估奠定基础。【方法】获取5个黑老虎个体的叶绿体基因组,通过基因注释,采用生物信息学方法开展核苷酸多态性、简单重复序列(simple sequarce repeat,SSR)分析,并以2个五味子属植物为外类群分析南五味子属的系统发育关系。【结果】黑老虎完整叶绿体基因组为典型的四分体环状结构,长度为145 413~145 903 bp。其中,大单拷贝区的长度范围为94 457~94 757 bp,小单拷贝区的长度为18 032~18 047 bp,重复区为16 431~16 552 bp。黑老虎叶绿体基因组编码125个基因,蛋白编码基因、tRNA基因、rRNA基因分别为82、35、8个;总G(鸟嘌呤)和C(胞嘧啶)的碱基数占总碱基数的比例(GC占比)为39.7%。采用滑动窗口法分析发现黑老虎叶绿体基因组上petN-psbM和trnS-GCU-trnG-UCC具有很高的核苷酸多态性(Pi > 0.03)。该物种叶绿体基因组上共有212个SSR位点,其中24个SSR位点在5个黑老虎个体间存在多态变异,具有应用前景。系统发育分析表明,此次研究中黑老虎个体聚成一支,与同属其他物种关系较远。【结论】本研究首次系统比较了多个黑老虎个体的叶绿体基因组,发掘出基因组内的高变异区域及变异SSR位点,为其遗传多样性及种质资源评估奠定了基础。
【Objective】This study aims to investigate the chloroplast genomes and SSR loci of Kadsura coccinea, an important plant resource in China, to establish a basis for assessing its genetic diversity and germplasm resources.【Method】The study obtained and analyzed the chloroplast genomes of five individuals of K. coccinea through genetical annotation, nucleotide polymorphism analysis, and SSR analysis using bioinformatics methods. Additionally, the phylogenetic analysis of Kadsura were reconstructed using two Schisandra spp. as outgroups.【Result】The chloroplast genome of K. coccinea showed a typical quadripartite structure, with genome lengths ranging from 145 413 to 145 903 base pairs (bp). The large single-copy region (LSC) spaned from 94 457 to 94 757 bp, while the small single-copy region (SSC) encompassed 18 032 to 18 047 bp. It encodes a total of 125 genes, including 82 protein-coding genes, 35 tRNA genes, and 8 rRNA genes. The genome had a total GC content of 39.7% and demonstrated substantial nucleotide polymorphisms (Pi > 0.03) in the intergenic regions of petN-psbM and trnS-GCU-trnG-UCC. A total of 212 single sequence repeat (SSR) loci were identified across the chloroplast genomes of this species. Mononucleotide repeats were the most prevalent, followed by trinucleotide repeats, and pentanucleotide repeats were the least frequent. Among these loci, 24 polymorphic SSR loci were found among five individuals of K. coccinea, indicating their potential utility in future. Phylogenetic analysis robustly clusters K. coccinea individuals into a distinct group, revealing no close relationship with other congeneric species.【Conclusion】This study presents the first systematic comparison of chloroplast genomes among multiple K. coccinea individuals. Our findings identified highly variable regions and SSR loci that can be valuable for evaluating the genetic diversity and germplasm resource of this species.
黑老虎 / 质体基因组 / 遗传多样性 / SSR位点 / 系统发育
Kadsura coccinea / plastid genome / genetic diversity / SSR loci / phylogeny
| [1] |
|
| [2] |
林祁, 段林东, 姚炳矾. 南五味子属(五味子科)三种植物之补记[J]. 植物分类学报, 2005, 43(6):567-570.
|
| [3] |
黄珊珊, 黄晓玲, 宋卉, 等. 中药黑老虎的研究进展[J]. 海峡药学, 2021, 33(11):38-40.
|
| [4] |
苏维, 王欣悦, 付港, 等. 南五味子属植物的化学成分、药理作用及临床应用研究进展[J]. 中国中药杂志, 2024, 49(1):26-38.
|
| [5] |
杨赛男, 戴斌, 潘清平, 等. 黑老虎植物资源利用研究进展[J]. 湖南生态科学学报, 2022, 9(3):112-120.
|
| [6] |
杨芝干. 地标奇果:通道“黑老虎”[J]. 生命世界, 2019(9):66-69.
|
| [7] |
林旭俊, 陆文, 李善志, 等. 药用植物黑老虎的资源调查[J]. 热带林业, 2019, 47(2):34-36.
|
| [8] |
韦霄, 梁惠凌, 唐辉, 等. 广西南五味子属植物的分布与利用[J]. 广西农业科学, 2006, 37(2):117-119.
|
| [9] |
邹建文, 罗先权, 饶红欣, 等. 常绿木质藤本植物黑老虎基因组SSR特征分析及引物开发[J]. 中南林业科技大学学报, 2021, 41(4):130-138.
|
| [10] |
|
| [11] |
|
| [12] |
赵儒楠, 褚晓洁, 刘维, 等. 鹅耳枥属树种叶绿体基因组结构及变异分析[J]. 南京林业大学学报(自然科学版), 2021, 45(2):25-34.
|
| [13] |
袁钰晨, 谢旭强, 徐立清, 等. 不同年龄阶段胡桃楸天然更新幼树的光合生理特性[J]. 森林工程, 2023, 39(4):29-37.
|
| [14] |
|
| [15] |
邓叶, 李翔, 李平, 等. 黑老虎种质资源与分子生物学研究进展[J]. 湖南生态科学学报, 2024, 11(1):96-104.
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
杨圆圆, 于世河, 卜鹏图, 等. 不同培育模式下日本落叶松林灌草和土壤养分特征研究[J]. 森林工程, 2023, 39(6):12-25.
|
| [35] |
|
| [36] |
|
| [37] |
刘玉壶. 木兰科[C]//中国科学院中国植物志编辑委员会. 中国植物志. 北京: 科学出版社, 1996.
|
| [38] |
|
| [39] |
毕海燕, 林祁, 刘长江, 等. 南五味子属(五味子科)的种子形态及其分类学意义[J]. 植物分类学报, 2002, 40(6):501-510.
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
/
| 〈 |
|
〉 |