 PDF(2727 KB)
PDF(2727 KB)


瓦雷兹芽孢杆菌FZB42中DUF1453家族蛋白CcdC的生物信息学分析及对菌株生长发育的影响
李美橘, 沈紫竹, 官陈郧, 樊奔, 赵银娟
南京林业大学学报(自然科学版) ›› 2025, Vol. 49 ›› Issue (6) : 247-254.
 PDF(2727 KB)
PDF(2727 KB)
 PDF(2727 KB)
PDF(2727 KB)
瓦雷兹芽孢杆菌FZB42中DUF1453家族蛋白CcdC的生物信息学分析及对菌株生长发育的影响
Bioinformatics analysis of the CcdC protein of the DUF1453 family in Bacillus velezensis FZB42 and its effects on growth and development
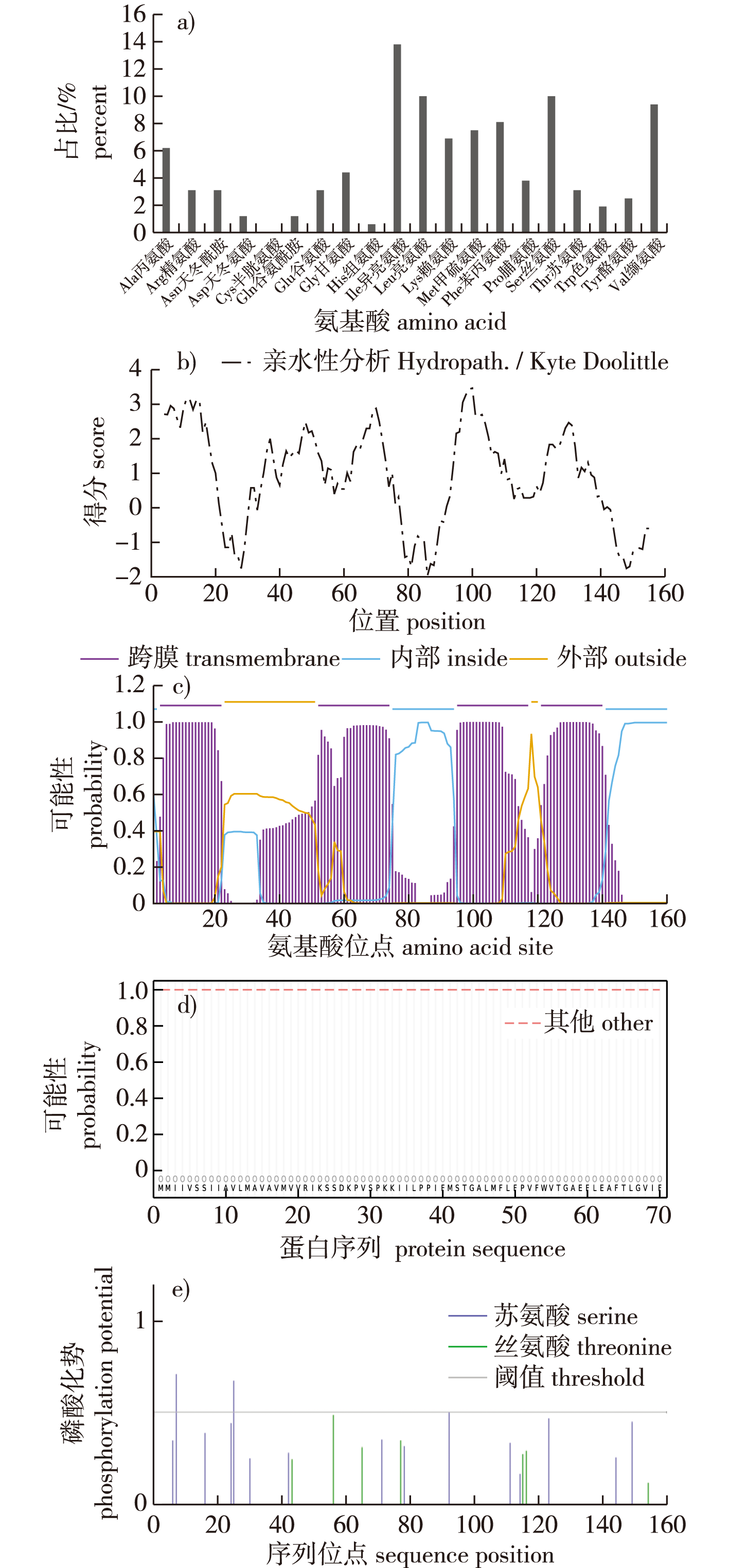
【目的】瓦雷兹芽孢杆菌(Bacillus velezensis) FZB42中的未知蛋白CcdC含有一个未知结构和功能的DUF1453结构域。利用生物信息学方法预测CcdC的生物学特性,并通过构建ccdC敲除株,研究CcdC蛋白的功能,为揭示DUF1453的结构和功能提供基础。【方法】利用生物信息学在线软件预测CcdC蛋白质的氨基酸组成及理化性质、疏水性、跨膜区、亚细胞定位等,并对不同细菌中的CcdC蛋白进行系统进化树分析。构建瓦雷兹芽孢杆菌FZB42的ccdC敲除株,比较野生型FZB42与ccdC敲除株在LB液体及固体培养基上的生长情况和DSM培养基中的芽孢形成情况。【结果】CcdC蛋白由160个氨基酸组成,它是一个位于细胞膜上且无信号肽的碱性、稳定、疏水的跨膜蛋白,存在3个丝氨酸磷酸化位点,其中第7位最有可能发生磷酸化。二级结构分析显示,CcdC蛋白主要以α-螺旋为主;三级结构分析显示,现有数据库中不存在与CcdC结构相似的蛋白质。通过菌落PCR,确定ccdC敲除株构建成功。比较FZB42与ccdC敲除株在生长和芽孢形成上的差异,发现ccdC基因的缺失影响了细菌生长和芽孢形成。【结论】生物信息学分析初步确定CcdC是一个膜蛋白,磷酸化位点的预测表明其可能参与信号传递功能,相互作用网络推测CcdC蛋白可能与生长发育有关。通过构建敲除菌株,明确了ccdC基因的缺失会影响细菌的生长发育,为后续探究DUF1453结构域的生物学功能和CcdC对瓦雷兹芽孢杆菌FZB42的生长发育调控奠定了基础。
【Objective】The CcdC protein in Bacillus velezensis FZB42 contains a DUF1453 domain with an unknown structure and function. This study aims to predict the biological properties of CcdC using bioinformatics methods and to investigate its function by constructing a ccdC knockout strain, thereby laying the groundwork for elucidating the structure and function of the DUF1453 domain.【Method】Bioinformatics online tools were employed to predict the amino acid composition, physicochemical properties, hydrophobicity, transmembrane regions, and subcellular localization of the CcdC protein. A phylogenetic tree analysis of CcdC proteins from different bacterial species was also conducted. A ccdC knockout strain of B. velezensis FZB42 was constructed, and the growth of the wild-type FZB42 strain and the ccdC knockout strain was compared in both liquid and solid LB media. Additionally, spore formation of both strains was assessed in DSM medium.【Result】The CcdC protein consisted of 160 amino acids and was a basic, stable, hydrophobic transmembrane protein located in the cell membrane, lacking a signal peptide. Secondary structure analysis revealed that the CcdC protein was predominantly α-helical. Tertiary structure analysis indicated that no structurally similar proteins to CcdC existed in current databases. Colony PCR confirmed the successful construction of the ccdC knockout strain. Comparative analysis of growth and spore formation between the wild-type FZB42 and the ccdC knockout strain demonstrated that the deletion of the ccdC gene significantly affected bacterial growth and spore formation.【Conclusion】Bioinformatics analysis tentatively identified CcdC as a membrane protein, with predicted phosphorylation sites suggesting its potential involvement in signal transduction functions. Interaction network analysis indicated that CcdC protein may be related to growth and development. The construction of the knockout strain confirmed that the deletion of the ccdC gene impair bacterial growth and development, providing a foundation for further research into the biological function of the DUF1453 domain and the regulatory role of CcdC in the growth and development of B. velezensis FZB42.
DUF1453结构域 / CcdC蛋白 / 生物信息学 / 生长发育 / 瓦雷兹芽孢杆菌
DUF1453 domain / CcdC protein / bioinformatics analysis / growth and development / Bacillus velezensis
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
An inherent problem in transmembrane protein topology prediction and signal peptide prediction is the high similarity between the hydrophobic regions of a transmembrane helix and that of a signal peptide, leading to cross-reaction between the two types of predictions. To improve predictions further, it is therefore important to make a predictor that aims to discriminate between the two classes. In addition, topology information can be gained when successfully predicting a signal peptide leading a transmembrane protein since it dictates that the N terminus of the mature protein must be on the non-cytoplasmic side of the membrane. Here, we present Phobius, a combined transmembrane protein topology and signal peptide predictor. The predictor is based on a hidden Markov model (HMM) that models the different sequence regions of a signal peptide and the different regions of a transmembrane protein in a series of interconnected states. Training was done on a newly assembled and curated dataset. Compared to TMHMM and SignalP, errors coming from cross-prediction between transmembrane segments and signal peptides were reduced substantially by Phobius. False classifications of signal peptides were reduced from 26.1% to 3.9% and false classifications of transmembrane helices were reduced from 19.0% to 7.7%. Phobius was applied to the proteomes of Homo sapiens and Escherichia coli. Here we also noted a drastic reduction of false classifications compared to TMHMM/SignalP, suggesting that Phobius is well suited for whole-genome annotation of signal peptides and transmembrane regions. The method is available at as well as at
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
孙柯, 赵学亮, 白丽艳, 等. 布鲁氏杆菌S2株DUF2326基因克隆与生物信息学分析[J]. 黑龙江畜牧兽医, 2020(5):9-14,148.
|
| [21] |
|
| [22] |
The DUF26 domain-containing protein is an extracellular structural protein, which plays an important role in signal transduction. Dongxiang wild rice ( Griff.) is the northern-most common wild rice in China. Using domain analysis, 85 DUF26 domain-containing genes were identified in Dongxiang wild rice (DXWR) and further divided into four categories. The DUF26 domain-containing genes were unevenly distributed on chromosomes, and there were 18 pairs of tandem repeats. Gene sequence analysis showed that there were significant differences in the gene structure and motif distribution of the DUF26 domain in different categories. Motifs 3, 8, 9, 13, 14, 16, and 18 were highly conserved in all categories. It was also found that there were eight plasmodesmata localization proteins (PDLPs) with a unique motif 19. Collinearity analysis showed that DXWR had a large number of orthologous genes with wheat, maize, sorghum and zizania, of which 17 DUF26 domain-containing genes were conserved in five gramineous crops. Under the stress of anaerobic germination and seedling submergence treatment, 33 DUF26 domain-containing genes were differentially expressed in varying degrees. Further correlation analysis with the expression of known submergence tolerance genes showed that these DUF26 domain-containing genes may jointly regulate the submergence tolerance process with these known submergence tolerance genes in DXWR.
|
| [23] |
Bacillus velezensis FZB42 is a plant growth-promoting rhizobacterium (PGPR) and a model microorganism for biofilm studies. Biofilms are required for the colonization and promotion of plant growth in the rhizosphere. However, little is known about how the final stage of the biofilm life cycle is regulated, when cells regain their motility and escape the mature biofilm to spread and colonize new niches. In this study, the non-annotated gene ccdC was found to be involved in the process of biofilm dispersion. We found that the ccdC-deficient strain maintained a wrinkled state at the late stage of biofilm formation in the liquid—gas interface culture, and the bottom solution showed a clear state, indicating that no bacterial cells actively escaped, which was further evidenced by the formation of a cellular ring (biofilm pellicle) located on top of the preformed biofilm. It can be concluded that dispersal, a biofilm property that relies on motility proficiency, is also positively affected by the unannotated gene ccdC. Furthermore, we found that the level of cyclic diguanylate (c-di-GMP) in the ccdC-deficient strain was significantly greater than that in the wild-type strain, suggesting that B. velezensis exhibits a similar mechanism by regulating the level of c-di-GMP, the master regulator of biofilm formation, dispersal, and cell motility, which controls the fitness of biofilms in Pseudomonas aeruginosain. In this study, we investigated the mechanism regulating biofilm dispersion in PGPR.
|
| [24] |
|
| [25] |
|
| [26] |
|
/
| 〈 |
|
〉 |