
- 国家林草科技领军期刊
- 中国精品科技期刊
- 中国高校百佳科技期刊
- 江苏省新闻出版政府奖期刊奖
- RCCSE林学权威期刊(A+)
- CSCD核心期刊
- Scopus数据库收录期刊
- 中文核心期刊
- SCD核心期刊

南京林业大学学报(自然科学版) ›› 2022, Vol. 46 ›› Issue (3): 143-150.doi: 10.12302/j.issn.1000-2006.202108023
收稿日期:2021-08-12
接受日期:2021-10-22
出版日期:2022-05-30
发布日期:2022-06-10
基金资助:Received:2021-08-12
Accepted:2021-10-22
Online:2022-05-30
Published:2022-06-10
摘要:
【目的】报道柳树痂囊腔菌(Elsinoë murrayae)的全基因组序列,与甜橙痂囊腔菌杨树致病型的基因组进行比较分析,为阐述柳树痂囊腔菌的致病和适应性机制提供参考。【方法】采用Illumina HiSeq 2500 测序仪对柳树痂囊腔菌的全基因组序列进行测序,预测蛋白编码基因,筛选与致病相关的碳水化合物活性酶基因、小分泌蛋白基因和次生代谢产物基因簇。根据痂囊腔属真菌基因的直系同源关系,筛选柳树痂囊腔菌和甜橙痂囊腔菌杨树致病型之间共有特异性的基因和二者之间差异基因,并进行GO富集分析。鉴定柳树痂囊腔菌的交配类型位点,使用特异性引物进行PCR,检测分离株的交配类型。【结果】组装获得了1个20.7 Mb基因组,完整度99%;预测出8 256个蛋白编码基因,其中包括486个碳水化合物活性酶基因,193个小分泌蛋白基因和16个次生代谢产物基因簇 (GenBank登录号:NKHZ00000000)。系统进化和共线性分析显示柳树痂囊腔菌和甜橙痂囊腔菌杨树致病型亲缘关系最近,两者之间具有12个在其他痂囊腔菌中没有的共有特异性基因。两个真菌的比较基因组分析,筛选出752和1 746个差异基因,主要参与碳水化合物代谢和毒素代谢的生物学过程。已有分离株的交配类型均为MAT1-2。【结论】获得柳树病原真菌-柳树痂囊腔菌的基因组,筛选出痂囊腔菌中负责寄主适应性的候选基因,分析了柳树痂囊腔菌交配系统,这可为柳树病害防治和柳树-病原真菌相互作用研究提供关键信息。
中图分类号:
程强,赵丽娟. 柳树痂囊腔菌的基因组测序和比较基因组分析[J]. 南京林业大学学报(自然科学版), 2022, 46(3): 143-150.
CHENG Qiang, ZHAO Lijuan. Draft genomes sequence of Elsinoë murrayae and comparative genomic analysis[J].Journal of Nanjing Forestry University (Natural Science Edition), 2022, 46(3): 143-150.DOI: 10.12302/j.issn.1000-2006.202108023.
表1
柳树痂囊腔菌(Emu)NL1菌株基因组组装注释统计"
| 参数features | 值 value |
|---|---|
| 测序总长/Gb total sequencing length | 6.3 |
| 总读序/Mb total reads | 42 |
| 覆盖度/倍 coverage | 241 |
| 基因组大小/bp genome size | 20 718 688 |
| scaffold数量 No. of scaffolds | 98 |
| GC含量/% GC content | 54.49 |
| N50/bp | 598 889 |
| 最大scaffold/bp the largest scaffold | 1 241 890 |
| 完整度/% completeness | 99 |
| 基因总数量 total genes | 8 317 |
| 蛋白编码基因数量 protein-coding genes | 8 256 |
| RNA编码基因数量 RNA genes | 66 |
| GenBank登录号 GenBank accession No. | NKHZ00000000 |
图1
Emu NL1致病相关蛋白分析 A.碳水化合物活性酶CAZymes;B.小分泌蛋白的特异性the specificity of SSPs;C.E. murrayae NL1和E. fawcettii的痂囊腔菌素合成基因簇the biosynthetic gene clusters of elsinochrome in E. murrayae NL1 and E. fawcettii。 箭头代表基因与转录方向,相同颜色箭头代表同源基因Arrows represent genes and their transcription directions, and arrows with the same color represent homologous genes。 CAC42_918/ELSFAW08003. 聚酮合成酶polyketide synthase; CAC42_919/ELSFAW08004.依赖FAD的单加氧酶FAD-dependent monooxygenase;CAC42_920/ELSFAW08005.依赖FAD/FMN的氧化还原酶FAD/FMN-dependent oxidoreductase;CAC42_921/ELSFAW08006. MFS转运体MFS transporter;CAC42_922/ELSFAW08007. O-甲基转移酶O-methyltransferase;CAC42_923/ELSFAW08008.锌指转录因子zing-finger transcription factor;CAC42_924/ELSFAW08009. MFS转运体MFS transporter;CAC42_925/ELSFAW08010.依赖氧或FAD/FMN的氧化还原酶oxygen-,FAD/FMN-dependent oxidoreductase;CAC42_926/ELSFAW08011.漆酶laccase;CAC42_927/ELSFAW08012, beta-ig-h3成束蛋白beta-ig-h3 fasciclin。"
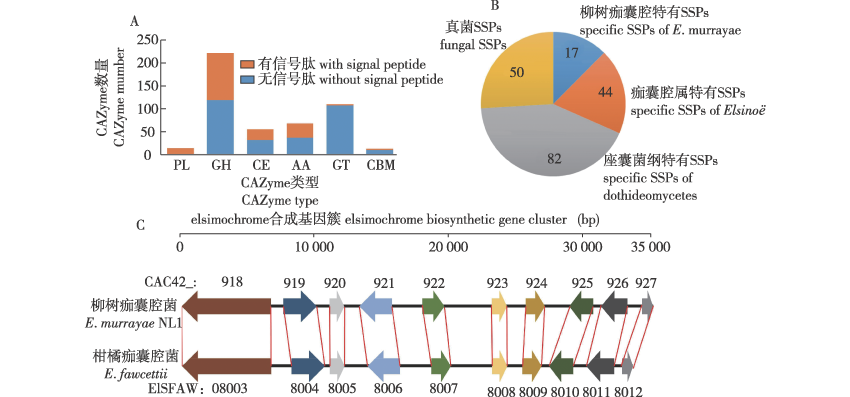
表2
柳树痂囊腔菌 NL1与痂囊腔属和多腔菌属 真菌的共线性分析"
| 真菌 fungi | 共线性 长度/Mb the aligned sequence size | 平均 一致性/% the average identity | 共线性 基因数量 the aligned gene No. |
|---|---|---|---|
| EauPSA NL1 | 19.86 | 74.5 | 6 561 |
| EauSOS Ea-1 | 19.55 | 74.4 | 6 278 |
| E. ampelina YL-1 | 19.50 | 73.21 | 6 239 |
| E. fawcettii SM16-1 | 19.83 | 73.21 | 6 234 |
| E. fawcettii DAR-70024 | 19.84 | 73.19 | 6 298 |
| E. fawcettii53147a | 19.44 | 73.13 | 6 448 |
| Myriangium duriaei | 17.01 | 70.14 | 3 854 |
表3
Emu致病相关蛋白"
| 类型 type | 蛋白编码基因位点编号 protein coding gene ID | 备注 note |
|---|---|---|
| Emu特有候选效应因子 specific candidate effectors | CAC42_3192, CAC42_8023, CAC42_2167, CAC42_1169, CAC42_2412, CAC42_1591, CAC42_5276, CAC42_5678, CAC42_4451, CAC42_1857, CAC42_1930, CAC42_1933, CAC42_7685, CAC42_1560, CAC42_3502, CAC42_4601, CAC42_1000 | 在公共数据中无同源蛋白 |
| Emu NL1与 Eau NL1(PSA) 共有特异性蛋白 the common specific proteins | CAC42_1526/B9Z65_5383,CAC42_1565/B9Z65_5438,CAC42_4946/B9Z65_8048,CAC42_5245/B9Z65_1324,CAC42_5855/B9Z65_7325,CAC42_6081/B9Z65_2782,CAC42_6597/B9Z65_8691,CAC42_7074/B9Z65_8229,CAC42_7195/B9Z65_4792,CAC42_7243/B9Z65_1695,CAC42_812/B9Z65_6144,CAC42_817/B9Z65_6139 | 只在E. murrayae 和E. australis中存在,而在Elsinoë属其他真菌中不存在的蛋白 |
| Emu NL1编码的差异蛋白中的小分泌蛋白 the small secreted proteins | CAC42_1169, CAC42_3924, CAC42_6199, CAC42_1000, CAC42_1442, CAC42_1560, CAC42_1591, CAC42_167, CAC42_1716, CAC42_1857, CAC42_1930, CAC42_1933, CAC42_1940, CAC42_1950, CAC42_2080, CAC42_2167, CAC42_2188, CAC42_2412, CAC42_2777, CAC42_3191, CAC42_3192, CAC42_3469, CAC42_3502, CAC42_4233, CAC42_4451, CAC42_4601, CAC42_4934, CAC42_5276, CAC42_5346, CAC42_5678, CAC42_5688, CAC42_569, CAC42_5736, CAC42_5768, CAC42_6556, CAC42_6573, CAC42_673, CAC42_6843, CAC42_7315, CAC42_7685, CAC42_8023, CAC42_8203 | E. murrayae NL1与E. australis NL1比对,所发现的差异蛋白,其中42个为小分泌蛋白 |
| [1] |
DICKMANN D I. Silviculture and biology of short-rotation woody crops in temperate regions: then and now[J]. Biomass Bioenergy, 2006, 30(8/9):696-705.DOI: 10.1016/j.biombioe.2005.02.008.
doi: 10.1016/j.biombioe.2005.02.008 |
| [2] |
ZHAO P, KAKISHIMA M, WANG Q, et al. Resolving the Melampsora epitea complex[J]. Mycologia, 2017, 109(3):391-407.DOI: 10.1080/00275514.2017.1326791.
doi: 10.1080/00275514.2017.1326791 |
| [3] |
WANG Y L, LU Q, JIA X Z, et al. First report of branch canker caused by Cytospora atrocirrhata on Populus sp.and Salix sp.in China[J]. Plant Dis, 2013, 97(3):426.DOI: 10.1094/PDIS-09-12-0854-PDN.
doi: 10.1094/PDIS-09-12-0854-PDN |
| [4] |
AYLWARD J, STEENKAMP E T, DREYER L L, et al. A plant pathology perspective of fungal genome sequencing[J]. IMA Fungus, 2017, 8(1):1-15.DOI: 10.5598/imafungus.2017.08.01.01.
doi: 10.5598/imafungus.2017.08.01.01 |
| [5] |
BUTIN H, KEHR R. Sphaceloma murrayae Jenk.& Grods.,a pathogen new to Europe on Salix spp.[J]. Forest Pathol, 2004, 34(1):27-31.DOI: 10.1046/j.1439-0329.2003.00344.x.
doi: 10.1046/j.1439-0329.2003.00344.x. |
| [6] |
SPIERS A G, HOPCROFT D H. Some electron microscope observations of conidium ontogeny of Sphaceloma murrayae on Salix[J]. N Z J Bot, 1992, 30(3):353-358.DOI: 10.1080/0028825X.1992.10412912.
doi: 10.1080/0028825X.1992.10412912 |
| [7] |
ZHAO L J, ZHANG W T, XIAO H J, et al. Molecular identification and characterization of Elsinoë murrayae (Synonym:Sphaceloma murrayae) from weeping willow[J]. J Phytopathol, 2018, 166(2):143-149.DOI: 10.1111/jph.12670.
doi: 10.1111/jph.12670 |
| [8] |
ZHAO L J, XIAO H J, MA X J, et al. Elsinoë australis causing spot anthracnose on poplar in China[J]. Plant Dis, 2020, 104(8):2202-2209.DOI: 10.1094/pdis-11-19-2349-re.
doi: 10.1094/pdis-11-19-2349-re |
| [9] |
LUO R B, LIU B H, XIE Y L, et al. SOAPdenovo2:an empirically improved memory-efficient short-read de novo assembler[J]. GigaScience, 2012, 1(1):18.DOI: 10.1186/2047-217X-1-18.
doi: 10.1186/2047-217X-1-18 |
| [10] |
WATERHOUSE R M, SEPPEY M, SIMÃO F A, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics[J]. Mol Biol Evol, 2018, 35(3):543-548.DOI: 10.1093/molbev/msx319.
doi: 10.1093/molbev/msx319 |
| [11] |
TER-HOVHANNISYAN V, LOMSADZE A, CHERNOFF Y O, et al. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training[J]. Genome Res, 2008, 18(12):1979-1990.DOI: 10.1101/gr.081612.108.
doi: 10.1101/gr.081612.108 |
| [12] |
LAGESEN K, HALLIN P, RØDLAND E A, et al. RNAmmer:consistent and rapid annotation of ribosomal RNA genes[J]. Nucleic Acids Res, 2007, 35(9):3100-3108.DOI: 10.1093/nar/gkm160.
doi: 10.1093/nar/gkm160 |
| [13] |
LOWE T M, CHAN P P. tRNAscan-SE on-line:integrating search and context for analysis of transfer RNA genes[J]. Nucleic Acids Res, 2016, 44(1):54-57.DOI: 10.1093/nar/gkw413.
doi: 10.1093/nar/gkw413 |
| [14] |
ZHANG H, YOHE T, HUANG L, et al. dbCAN2:a meta server for automated carbohydrate-active enzyme annotation[J]. Nucleic Acids Res, 2018, 46(1):95-101.DOI: 10.1093/nar/gky418.
doi: 10.1093/nar/gky418 |
| [15] |
ALMAGRO ARMENTEROS J J, TSIRIGOS K D, SØNDERBY C K, et al. Signal P 5.0 improves signal peptide predictions using deep neural networks[J]. Nat Biotechnol, 2019, 37(4):420-423.DOI: 10.1038/s41587-019-0036-z.
doi: 10.1038/s41587-019-0036-z |
| [16] |
KROGH A, LARSSON B, VON HEIJNE G, et al. Predicting transmembrane protein topology with a hidden Markov model:application to complete genomes[J]. J Mol Biol, 2001, 305(3):567-580.DOI: 10.1006/jmbi.2000.4315.
doi: 10.1006/jmbi.2000.4315 |
| [17] |
BLIN K, SHAW S, STEINKE K, et al. antiSMASH 5.0:updates to the secondary metabolite genome mining pipeline[J]. Nucleic Acids Res, 2019, 47(1):81-87.DOI: 10.1093/nar/gkz310.
doi: 10.1093/nar/gkz310 |
| [18] |
KUMAR S, STECHER G, TAMURA K. MEGA7:molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7):1870-1874.DOI: 10.1093/molbev/msw054.
doi: 10.1093/molbev/msw054 |
| [19] |
CASTILLO A I, NELSON A D L, HAUG-BALTZELL A K, et al. A tutorial of diverse genome analysis tools found in the CoGe web-platform using Plasmodium spp.as a model[J]. Database (Oxford), 2018,2018(10.1093):database.DOI: 10.1093/database/bay030.
doi: 10.1093/database/bay030 |
| [20] |
LI L, STOECKERT C J, ROOS D S. OrthoMCL: identification of ortholog groups for eukaryotic genomes[J]. Genome Res, 2003, 13(9):2178-2189. DOI: 10.1101/gr.1224503.
doi: 10.1101/gr.1224503 |
| [21] |
CHEN C J, CHEN H, ZHANG Y, et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8):1194-1202.DOI: 10.1016/j.molp.2020.06.009.
doi: 10.1016/j.molp.2020.06.009 |
| [22] |
STERGIOPOULOS I, DE WIT P J. Fungal effector proteins[J]. Annu Rev Phytopathol, 2009, 47:233-263.DOI: 10.1146/annurev.phyto.112408.132637.
doi: 10.1146/annurev.phyto.112408.132637 |
| [23] |
EBERT M K, SPANNER R E, DE JONGE R, et al. Gene cluster conservation identifies melanin and perylenequinone biosynthesis pathways in multiple plant pathogenic fungi[J]. Environ Microbiol, 2019, 21(3):913-927.DOI: 10.1111/1462-2920.14475.
doi: 10.1111/1462-2920.14475 |
| [24] |
LI Z, FAN Y C, CHANG P P, et al. Genome sequence resource for Elsinoë ampelina,the causal organism of grapevine anthracnose[J]. Mol Plant Microbe Interact, 2020, 33(4):576-579.DOI: 10.1094/MPMI-12-19-0337-A.
doi: 10.1094/MPMI-12-19-0337-A |
| [25] |
JEFFRESS S, ARUN-CHINNAPPA K, STODART B, et al. Genome mining of the Citrus pathogen Elsinoë fawcettii;prediction and prioritisation of candidate effectors,cell wall degrading enzymes and secondary metabolite gene clusters[J]. PLoS One, 2020, 15(5):e0227396.DOI: 10.1371/journal.pone.0227396.
doi: 10.1371/journal.pone.0227396 |
| [26] |
SHANMUGAM G, JEON J, HYUN J W. Draft genome sequences of Elsinoë fawcettii and Elsinoë australis causing scab diseases on Citrus[J]. Mol Plant Microbe Interactions, 2020, 33(2):135-137.DOI: 10.1094/mpmi-06-19-0169-a.
doi: 10.1094/mpmi-06-19-0169-a |
| [27] |
FAN X L, BARRETO R W, GROENEWALD J Z, et al. Phylogeny and taxonomy of the scab and spot anthracnose fungus Elsinoë (Myriangiales,Dothideomycetes)[J]. Stud Mycol, 2017, 87:1-41.DOI: 10.1016/j.simyco.2017.02.001.
doi: 10.1016/j.simyco.2017.02.001 |
| [28] |
NI M, FERETZAKI M, SUN S, et al. Sex in fungi[J]. Annu Rev Genet, 2011, 45:405-430.DOI: 10.1146/annurev-genet-110410-132536.
doi: 10.1146/annurev-genet-110410-132536 |
| [29] |
WILKEN P M, STEENKAMP E T, WINGFIELD M J, et al. Which MAT gene? Pezizomycotina (Ascomycota) mating-type gene nomenclature reconsidered[J]. Fungal Biol Rev, 2017, 31(4):199-211.DOI: 10.1016/j.fbr.2017.05.003.
doi: 10.1016/j.fbr.2017.05.003 |
| [30] |
CHUNG K R. Elsinoë fawcettii and Elsinoë australis:the fungal pathogens causing Citrus scab[J]. Mol Plant Pathol, 2011, 12(2):123-135.DOI: 10.1111/j.1364-3703.2010.00663.x.
doi: 10.1111/j.1364-3703.2010.00663.x. |
| [1] | 邓演文, 田淑意, 刘世晗, 梁建, 陶家璐, 邓小梅. 广东含笑和诗琳通含笑叶绿体基因组结构及系统发育分析[J]. 南京林业大学学报(自然科学版), 2025, 49(1): 69-77. |
| [2] | 欧阳泽怡, 李志辉, 牟虹霖, 姜小龙, 程勇, 吴际友. 基于简化基因组开发青冈和滇青冈微卫星引物[J]. 南京林业大学学报(自然科学版), 2024, 48(6): 62-70. |
| [3] | 翟学昌, 彭丽, 颜海飞, 朱柯帆, 张淑燕, 张彩云, 鲁显楷. 资源植物黑老虎的比较叶绿体基因组学研究[J]. 南京林业大学学报(自然科学版), 2024, 48(6): 71-78. |
| [4] | 马建慧, 陈昕, 耿礼阳, 汤晨茜, 魏雪妍. 蔷薇科花楸属白毛系的系统发生分析[J]. 南京林业大学学报(自然科学版), 2024, 48(4): 25-36. |
| [5] | 刘莉, 瞿印权, 余延浩, 王倩, 洑香香. 青钱柳全基因组SSR位点分析及多态性引物开发[J]. 南京林业大学学报(自然科学版), 2024, 48(4): 67-75. |
| [6] | 杨永. 裸子植物的系统分类:历史、现状和展望[J]. 南京林业大学学报(自然科学版), 2024, 48(3): 14-26. |
| [7] | 尹增芳, 欧香, 陈瑶, 杨爱香, 孙李勇. 望春玉兰生物学基础研究进展与展望[J]. 南京林业大学学报(自然科学版), 2024, 48(2): 256-262. |
| [8] | 邱靖, 李嘉宝, 朱大海, 陈昕. 花楸属直脉组7种/变种基因组大小及叶表皮微形态特征的分类学意义[J]. 南京林业大学学报(自然科学版), 2023, 47(3): 77-86. |
| [9] | 马秋月, 王玉虓, 李倩中, 李淑顺, 闻婧, 朱璐, 颜坤元, 杜一鸣, 解志军, 李淑娴, 欧阳芳群, 鲁成代. 基于流式细胞术和K-mer方法测定6种槭属植物基因组大小[J]. 南京林业大学学报(自然科学版), 2023, 47(1): 163-170. |
| [10] | 陈赢男, 韦素云, 曲冠正, 胡建军, 王军辉, 尹佟明, 潘惠新, 卢孟柱, 康向阳, 李来庚, 黄敏仁, 王明庥. 现代林木育种关键核心技术研究现状与展望[J]. 南京林业大学学报(自然科学版), 2022, 46(6): 1-9. |
| [11] | 储陈辰, 孙明升, 吴雨涵, 言震宇, 李婷, 冯洋帆, 国颖, 尹佟明, 薛良交. 杨树泛基因组构建与基因组变异分析[J]. 南京林业大学学报(自然科学版), 2022, 46(6): 251-260. |
| [12] | 何旭东, 隋德宗, 王红玲, 黄瑞芳, 郑纪伟, 王保松. 中国柳树遗传育种研究进展[J]. 南京林业大学学报(自然科学版), 2022, 46(6): 51-63. |
| [13] | 方炎明, 朱福远, 李垚, 李璇. 基于基因组学的栎树生物学研究进展[J]. 南京林业大学学报(自然科学版), 2022, 46(6): 64-72. |
| [14] | 王子玥, 甄艳, 刘光欣, 席梦利. 染色质转座酶可及性测序及其在木本植物中的应用前景[J]. 南京林业大学学报(自然科学版), 2022, 46(5): 1-10. |
| [15] | 丁晓磊, 张悦, 林司曦, 叶建仁. 基于高通量测序技术的松材线虫研究进展[J]. 南京林业大学学报(自然科学版), 2022, 46(4): 1-7. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
