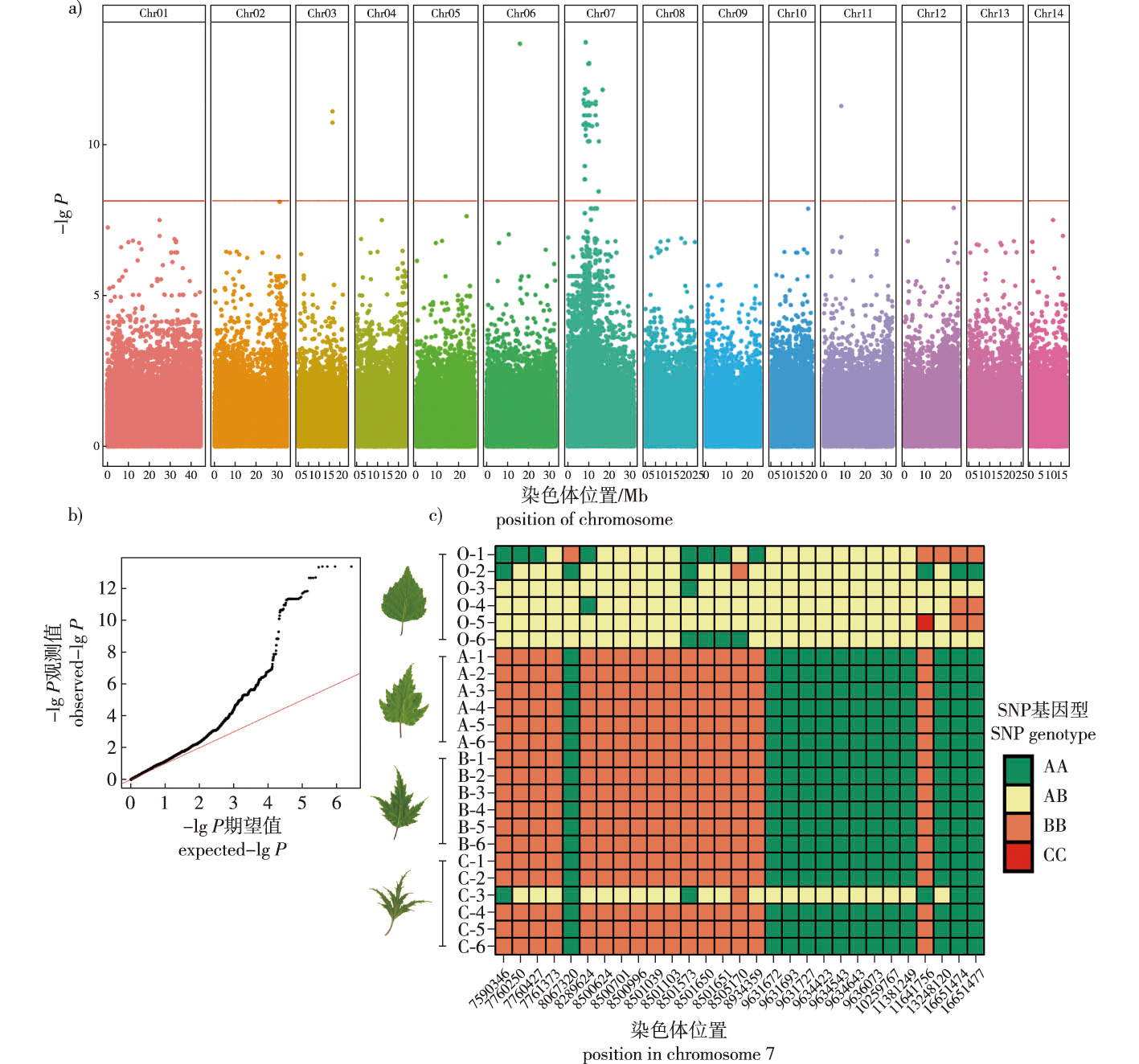
【Objective】 As a variety of Betula pendula, B. pendula ‘Dalecarlica’ exhibits lobed shaped split leaves, which have significant application value in horticultural ornamental, breeding, and plant developmental biology research. Understanding the mechanisms of leaf shape variation in B. pendula ‘Dalecarlica’ has academic value in the field of genetics, and identifying the genes corresponding to this trait is important for further research on this topic. This study aimed to preliminarily locate and identify the genes regulating the leaf shape of B. pendula ‘Dalecarlica’, and screen and annotate potential candidate genes, thus laying the groundwork for subsequent validation experiments and mechanistic research. 【Method】 A total of 24 offspring individuals from a single B. pendula ‘Dalecarlica’ maternal tree were selected as experimental materials. This half-sibling family included the offspring of individuals with different leaf types, including normal, week-lobed, lobed and strong-lobed leaves. Five leaves were collected from each plant and scanned. Leaf shape irregularity degree (Dli) was measured as Dli=4πS/C2, and which for each individual was calculated. The significance of Dli differences among individuals was analyzed. RNA-Seq data were obtained for each individual, and SNP/InDel information was called. Using the degree of leaf shape irregularity as a phenotype, an association analysis was performed using a generalized linear model (GLM). The association area were delineated based on SNPs/InDels with significant association signals. Furthermore, gene expression levels of the entire genome in the experimental materials were analyzed based on RNA-Seq data. The significance of gene transcription level differences was calculated, and differentially expressed genes (DEGs) within the association area were identified. SNPs/InDels with association P values smaller than the Bonferroni threshold were considered associated SNPs/InDels. Genes affected by associated SNPs/InDels and differentially expressed genes were considered candidate genes. Gene functional annotations were performed using online databases. 【Result】 The degree of leaf irregularity showed extremely significant differences among different individuals within the experimental population (P < 0.01). The coefficient of variation for leaf shape irregularity in the experimental population was 60.51%, which met the requirements for association analysis. A total of 331 Gb of RNA-Seq data was generated, of which 3 303 530 SNPs/InDels were detected. After filtering, 1 367 359 high-quality polymorphic SNPs/InDels were retained. Using the degree of leaf shape irregularity as the phenotype, association analysis was performed on these SNPs/InDels using a GLM. Association analysis identified 66 SNPs/InDels with significant P values below the Bonferroni threshold. By closely linking these 61 SNPs/InDels, the association area containing the leaf shape-regulating genes of B. pendula ‘Dalecarlica’ was defined, spanning a length of 9.18 MbP and containing 400 genes. Within this area, 61 associated SNPs/InDels were found to affect the coding of 17 known genes. Analysis of gene expression levels revealed that of the 400 genes within the association area, 25 genes showed extremely significant differences (P < 0.01) between B. pendula, B. pendula ‘Dalecarlica’. Functional candidate gene annotation identified three transcription factors, three proteins involved in cell composition, six proteins involved in protein translation processing, nine protein kinases involved in signal transduction, five enzymes involved in biochemical reactions, three proteins involved in substance transport, one protein involved in the post-transcriptional modification of RNA, one DNA endonuclease/exonuclease, and one protein involved in stomatal regulation. Additionally, eight genes could not be found in the existing online databases, and their types and functions remain unknown. Finally, three transcription factors and one auxin carrier protein were considered as possible key genes regulating the leaf shape of B. pendula ‘Dalecarlica’. 【Conclusion】 The gene locus responsible for regulating the splitting trait of B. pendula ‘Dalecarlica’ leaves was mapped to the interval of chromosome 7:7466344-16651477, with a total of 40 candidate genes located within this association area. Based on the gene annotation results, future studies should focus on the three identified transcription factors and one auxin carrier protein.
 PDF(2557 KB)
PDF(2557 KB)


 PDF(2557 KB)
PDF(2557 KB)
 PDF(2557 KB)
PDF(2557 KB)