 PDF(2262 KB)
PDF(2262 KB)


Establishment and application of a flow cytometry method for chromosome ploidy identification of Cyclocarya paliurus
SONG Ziqi, BIAN Guoliang, LIN Feng, HU Fengrong, SHANG Xulan
Journal of Nanjing Forestry University (Natural Sciences Edition) ›› 2024, Vol. 48 ›› Issue (2) : 61-68.
 PDF(2262 KB)
PDF(2262 KB)
 PDF(2262 KB)
PDF(2262 KB)
Establishment and application of a flow cytometry method for chromosome ploidy identification of Cyclocarya paliurus
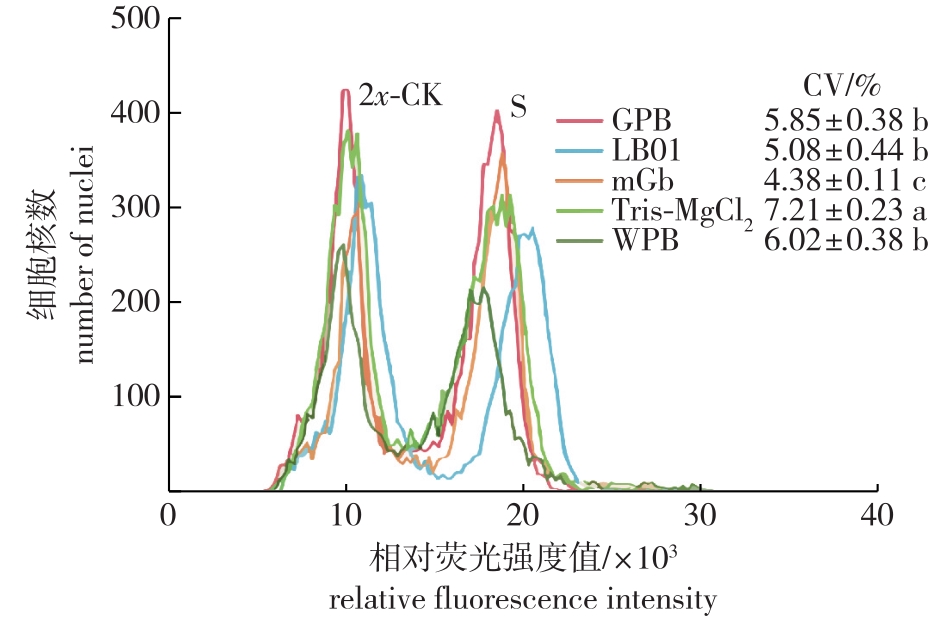
【Objective】 To provide technical support and basic data for the germplasm identification and genetic breeding of Cyclocarya paliurus, this study determined the chromosome ploidy using flow cytometry. 【Method】 C. paliurus leaves were used as study materials to compare the ploidy detection effects of different nuclei isolation buffers, centrifugation treatments, and leaf preservation methods. The ploidy identification method for C. paliurus using flow cytometry was as follows: 0.50-1.00 cm2 leaves of the reference sample and the test sample were mixed and chopped with 1 mL of mGb buffer. After filtration, 20 μL PI was added for staining for 1 min. The ploidy of 1 395 C. paliurus germplasm resources was determined by the established method. 【Result】 The optimal choice for isolating nuclei and achieving a clear peak in the resulting nucleus suspension was the use of Modified Gitschier buffer (mGb). Nuclear suspensions could be directly stained after filtration without centrifugation treatment. The optimal detection effect was obtained for leaves preserved at 4 ℃, and the most suitable storage time was 7 d. Drying leaves with silica gel yielded superior test results compared to freezing them, and the most suitable storage time for silica gel drying was 150 d. The coefficients of variation of 100 test samples ranged from 2.13% to 5.04%. If the estimated value of ploidy was 1.80-2.40, the sample was identified as diploid. If the estimated value of ploidy was 3.60-4.20, the sample was identified as tetraploid. When the estimated value of ploidy was 3.00 ± 0.40, the reference sample with the same ploidy as the initial judgment was used for the second detection. Using this method for the ploidy identification of 1 395 germplasm resources 104 diploids and 1 291 tetraploids were detected. 【Conclusion】 Samples with an abnormal estimated value of ploidy could be identified quickly by the second detection with the same ploidy standard. The method is simple, efficient, and accurate, and provides an effective method for ploidy identification of C. paliurus germplasm.
Cyclocarya paliurus / ploidy identification / flow cytometry / nuclei isolation buffer / internal standard method
| [1] |
方升佐. 青钱柳产业发展历程及资源培育研究进展[J]. 南京林业大学学报(自然科学版), 2022, 46(6): 115-126.
|
| [2] |
洪俊溪. 青钱柳人工林材性试验研究[J]. 福建林学院学报, 1997, 17(3): 214-217.
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
何法慧, 左倩倩, 于景金, 等. 35份狗牙根种质材料指纹图谱构建及染色体倍性鉴定[J]. 南京农业大学学报, 2023, 46(1):42-54.
|
| [10] |
赵帅琪, 张伟伟, 牛俊芳, 等. 森林草莓和栽培草莓在果实发育和成熟过程中细胞壁变化的比较[J]. 植物生理学报, 2021, 57(12): 2323-2336.
|
| [11] |
康向阳. 杜仲良种选育研究现状及展望[J]. 北京林业大学学报, 2017, 39(3): 1-6.
|
| [12] |
李秀兰, 陈力. 三倍体丹参的培育及其可持续利用研究[J]. 中草药, 2012, 43(2): 375-379.
|
| [13] |
|
| [14] |
|
| [15] |
陶抵辉, 李小红, 王利群, 等. 植物染色体倍性鉴定方法研究进展[J]. 生命科学研究, 2009, 13(5): 453-458.
|
| [16] |
|
| [17] |
|
| [18] |
金亮, 徐伟韦, 李小白, 等. DNA流式细胞术在植物遗传及育种中的应用[J]. 中国细胞生物学学报, 2016, 38(2): 225-234.
|
| [19] |
宫雅昕, 岳涵, 向宇, 等. GABA代谢负调控叶片细胞内复制发生的机制研究[J]. 植物生理学报, 2020, 56(2): 235-246.
|
| [20] |
|
| [21] |
田新民, 周香艳, 弓娜. 流式细胞术在植物学研究中的应用检测植物核DNA含量和倍性水平[J]. 中国农学通报, 2011, 27(9): 21-27.
|
| [22] |
韩杰, 沈海萍, 储冬生, 等. 4个薄壳山核桃品种核型分析[J]. 分子植物育种, 2018, 16(17): 5704-5711.
|
| [23] |
任伟超, 徐姣, 樊锐锋, 等. 应用流式细胞术对柳属染色体倍性与基因组大小测定[J]. 东北林业大学学报, 2021, 49(4): 56-61.
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
张桂芳, 王艳, 闫小巧, 等. 流式细胞仪检测铁皮石斛核DNA初探[J]. 现代中药研究与实践, 2017, 31(1): 16-19.
|
| [30] |
于红梅, 王静, 赵密珍, 等. 利用流式细胞仪检测草莓倍性方法的优化[J]. 南方农业学报, 2012, 43(10): 1530-1533.
|
| [31] |
杨静, 宋勤霞, 宁军权, 等. 利用流式细胞术鉴定桑树染色体倍性的方法[J]. 蚕业科学, 2017, 43(1): 8-17.
|
| [32] |
何婷, 郭桂梅, 陆瑞菊, 等. 两份大麦材料小孢子诱导愈伤及再生植株的倍性研究[J]. 植物生理学报, 2021, 57(8): 1708-1714.
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
李雯雯, 刘立强, 帕米尔·艾尼, 等. 利用流式细胞术鉴定新疆野杏染色体倍性和DNA含量[J]. 农业生物技术学报, 2019, 27(3): 542-550.
|
| [38] |
|
| [39] |
|
| [40] |
吕顺, 任毅, 王芳, 等. 利用流式细胞术快速鉴定169份香蕉种质资源的染色体倍性[J]. 果树学报, 2018, 35(6): 668-684.
|
/
| 〈 |
|
〉 |