 PDF(2779 KB)
PDF(2779 KB)


Analyzing the genetic diversity of Nymphaea spp. based on SSR markers
MAO Liyan, LI Huimin, LONG Lingyun, HUANG Qiuwei, TANG Yuwei, YU Yanping, HUANG Xinyi, TAN Xiaohui, NONG Xiaohui, ZHU Tianlong, LU Zushuang
Journal of Nanjing Forestry University (Natural Sciences Edition) ›› 2024, Vol. 48 ›› Issue (5) : 57-68.
 PDF(2779 KB)
PDF(2779 KB)
 PDF(2779 KB)
PDF(2779 KB)
Analyzing the genetic diversity of Nymphaea spp. based on SSR markers
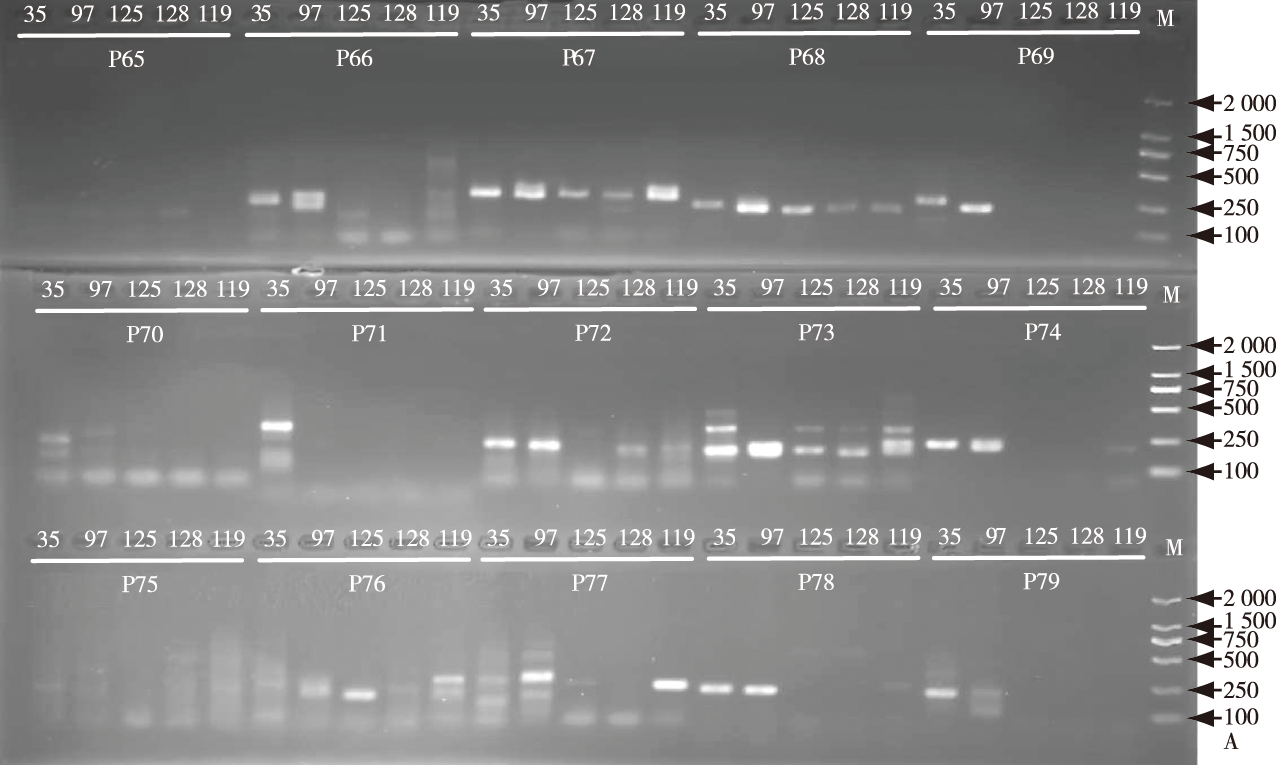
【Objective】Nymphaea spp. (waterlily) are important aquatic flowers. The rapid development of the waterlily industry has led to challenges in identifying genetic backgrounds of newly introduced germplasms in a timely manner due to inadequate introduction management practices. This has resulted in issues such as mislabeling of seedlings and unclear parentage, hampering the effective utilization and innovation of waterlily germplasm resources. This study focuses on developing genome-wide SSR markers for conducting phylogenetic and genetic diversity analyses of waterlily species. These markers are intended to serve as a theoretical foundation for conserving and breeding waterlily germplasm resources.【Method】Based on the published complete genome sequence of waterlily, the SSR loci in 14 chromosome genes were analyzed using the micro satellite identification (MISA) tool. Subsequently, 150 pairs of SSR primers were designed with the assistance of Primer 3.0 software. Five native germplasms were chosen for PCR amplification utilizing the 150 primer pairs. Following PCR, SSR primers demonstrating high polymorphism were identified through agarose and denaturing polyacrylamide gel electrophoresis. The selected SSR primers were synthesized with fluorescent primers (FAM and HEX) before amplifying 147 waterlily samples via PCR. The resulting products were then assessed using capillary electrophoresis, and the raw data were analyzed using GeneMarker software. Fragment sizes at each allele site for every sample were compiled into a 0/1 matrix. Genetic diversity indices, cluster analysis, and principal component analysis were computed using Popgenen and NTSYS software toos.【Result】11 pairs of SSR primers exhibiting distinct bands and high polymorphism were carefully chosen following the analysis of agarose gel and polyacrylamide gel electrophoresis results. These selected primers were employed to evaluate the genetic diversity among 147 waterlilies. Capillary electrophoresis revealed the presence of 307 alleles. The polymorphism index (PIC) ranged from 0.46 to 0.60, average at 0.53. The effective allele number (Ne) rangedfrom 1.042 8 to 1.117 5, with an average of 1.071 8. Nei’s gene diversity index (H) ranged from 0.038 0 to 0.086 2, averaging at 0.056 2. The Shannon diversity information index (I) ranged from 0.085 6 to 0.163 8, averaging at 0.114 4. Cluster analysis indicated genetic similarity coefficients ranging from 0.781 8 to 0.993 5 among the 147 waterlilies, with an average coefficiency of 0.899 2. These waterlilies were classified into six groups based on a genetic similarity coefficient was 0.879 0. Principal coordinates analysis (PCoA) displayed results closely aligned with traditional morphological classification for the 147 waterlilies. The first, second and third principal coordinates accounted for 16.54%, 8.35% and 5.43% of the total genetic variation, respectively, comprising 30.32% altogether. The first coordinate was correlated with aroma formation and stamen development, the second with flowering time and environmental adaptability, and the third with flower types.【Conclusion】11 pairs of SSR primers displaying substantial polymorphism were chosen, demonstrating efficacy in distinguishing the genetic relationships among 147 waterlilies. These waterlilies were systematically categorized into six primary branches, with classification outcomes mirroring traditional morphology categorizations. The selected 11 pairs of SSR primers have potential utility in analyzing genetic diversity and identifying waterlily species. The SSR markers findings stand poised to offer a robust scientific foundation for germplasm collection, preservation, innovation, and the development of waterlily species.
Nymphaea spp. (waterlily) / SSR molecular marker / genetic diversity / capillary electrophoresis
| [1] |
李淑娟, 尉倩, 陈尘, 等. 中国睡莲属植物育种研究进展[J]. 植物遗传资源学报, 2019, 20(4):829-835.
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
黄国振, 邓惠勤, 李祖修, 等. 睡莲[M]. 北京: 中国林业出版社, 2009:1-10.
|
| [8] |
万里波, 王奎玲, 刘庆华, 等. 青岛耐寒睡莲品种数量分类研究[C]// 青岛: 中国观赏园艺研究进展(2014), 2014:133-139.
|
| [9] |
苏群, 杨亚涵, 田敏, 等. 49份睡莲资源表型多样性分析及综合评价[J]. 西南农业学报, 2019, 32(11):2670-2681.
|
| [10] |
张海平. 部分睡莲属植物的形态多样性及同工酶分析[D]. 南京: 南京农业大学, 2008.
|
| [11] |
袁茹玉. 不同品种睡莲花挥发物组成及其茶汤功能成分和抗氧化活性评价[D]. 南京: 南京农业大学, 2014.
|
| [12] |
刘闵豪, 肖兴翠, 张春花, 等. 基于SSR的四川主要香椿种源遗传多样性分析[J]. 中南林业科技大学学报, 2022, 42(12):60-67.
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
苏群, 杨亚涵, 田敏, 等. 睡莲种质资源遗传多样性分析及DNA指纹图谱构建[J]. 热带作物学报, 2020, 41(2):258-266.
|
| [17] |
李志强, 吴超, 贺熙勇, 等. 基于SSR标记的澳洲坚果种质资源DNA指纹图谱的构建[J]. 果树学报, 2022, 39(11):2028-2035.
|
| [18] |
梁燕, 韩传明, 孙超, 等. 基于SSR标记的核桃种质资源遗传多样性与遗传结构分析[J]. 北方园艺, 2022(9):47-54.
|
| [19] |
蔡元保, 杨祥燕, 陈豪军, 等. SRAP结合SCoT标记分析番木瓜种质的遗传多样性[J]. 植物遗传资源学报, 2014, 15(2):292-298.
|
| [20] |
井敏敏, 黄炳钰, 戴小红, 等. 基于SSR标记的澳洲坚果种质资源遗传多样性分析[J]. 热带作物学报, 2022, 43(2):262-270.
|
| [21] |
任雪锋, 邓亚博, 臧国长, 等. 基于SSR标记的河南省狗牙根遗传多样性及群体遗传结构分析[J]. 草业学报, 2022, 31(3):60-70.
|
| [22] |
郑永胜, 张晗, 王东建, 等. 基于荧光检测技术的小麦品种SSR鉴定体系的建立[J]. 中国农业科学, 2014, 47(19):3725-3735.
|
| [23] |
苏群, 王虹妍, 卢家仕, 等. 睡莲的SSR引物对及合成方法和应用:CN113832254A[P]. 2021-12-24.
|
| [24] |
|
| [25] |
胡永超, 马洁, 唐建宁, 等. 不同树龄枸杞古树的遗传多样性研究[J]. 植物遗传资源学报, 2022, 23(3):755-767.
|
| [26] |
仲小茹, 柯叮, 黄献峰, 等. 基于SSR标记的江西省枫香古树遗传多样性评价[J]. 植物遗传资源学报, 2023, 24(2):523-531.
|
| [27] |
张旻桓, 姚奕平, 黄宇, 等. 基于SSR标记的江南牡丹品种群遗传多样性及亲缘关系[J]. 中南林业科技大学学报, 2023, 43(1):164-172.
|
| [28] |
孙春青, 陶美奇, 姚悦梅, 等. 睡莲生殖器官发育过程中解剖结构的变化[J]. 植物资源与环境学报, 2022, 31(1):21-28.
|
| [29] |
孙春青, 潘跃平, 单延博, 等. 睡莲品种墨宝自交结实率低的细胞学机理[J]. 江苏农业学报, 2017, 33(4):890-894.
|
| [30] |
黄祥, 杨梅花, 楚光明, 等. 耐寒睡莲种质资源分析及观赏性评价[J]. 分子植物育种, 2022, 20(4):1348-1357.
|
| [31] |
潘庆龙, 付瑛格, 谷佳, 等. 海南引种睡莲表型多样性分析及评价[J]. 热带作物学报, 2021, 42(10):2777-2788.
|
| [32] |
张大乐, 李锁平, 雷进生, 等. 利用SSR标记对12个啤酒大麦品种的聚类分析和主坐标分析[J]. 河北农业大学学报, 2007, 30(3):26-31.
|
| [33] |
赵亚军, 王灏, 穆建新, 等. 油菜自育与其他主栽品种的遗传多样性和遗传关系分析[J]. 分子植物育种, 2018, 16(8):2714-2722.
|
| [34] |
肖志娟, 翟梅枝, 王振元, 等. 微卫星DNA在分析核桃遗传多样性上的应用[J]. 中南林业科技大学学报, 2014, 34(2):55-61.
|
/
| 〈 |
|
〉 |