 PDF(2051 KB)
PDF(2051 KB)


Genome wide SSR analysis and primer development of Osmanthus fragrans
ZHU Yue, ZHANG Min, ZHANG Cheng, MA Cancan, YAN Jiping, DUAN Yifan, WANG Xianrong
Journal of Nanjing Forestry University (Natural Sciences Edition) ›› 2025, Vol. 49 ›› Issue (5) : 174-182.
 PDF(2051 KB)
PDF(2051 KB)
 PDF(2051 KB)
PDF(2051 KB)
Genome wide SSR analysis and primer development of Osmanthus fragrans
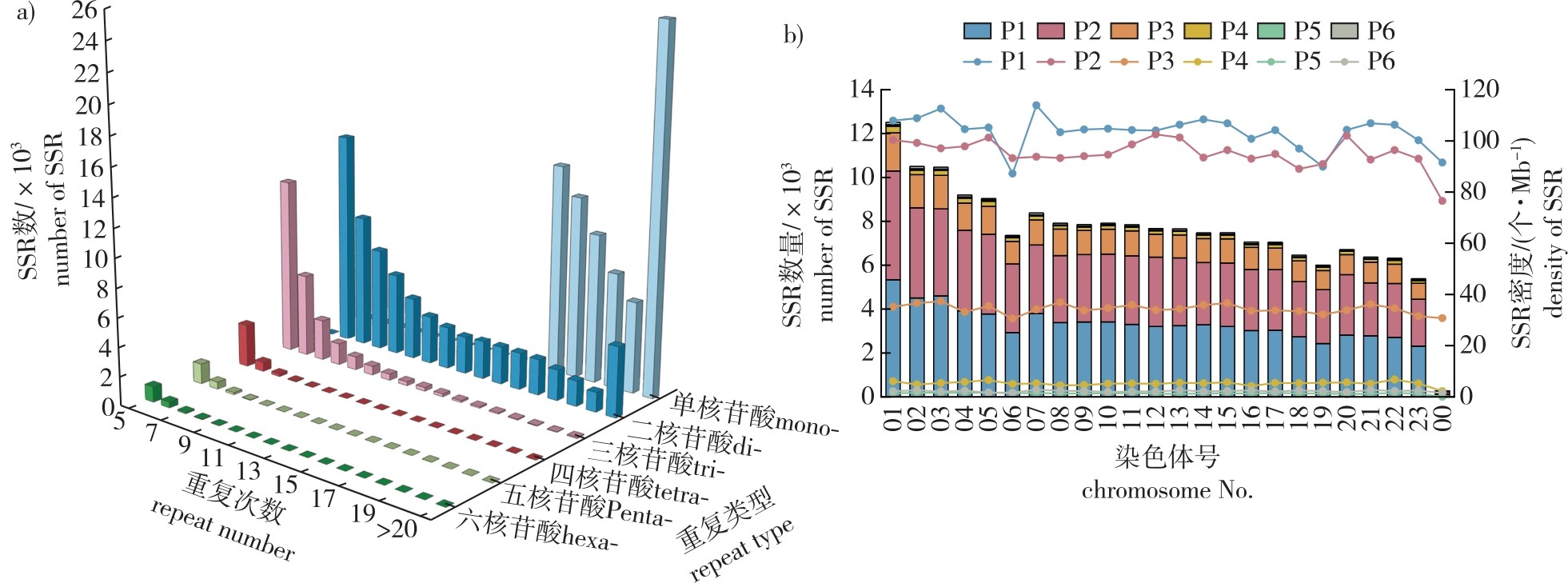
【Objective】Osmanthus fragrans is one of the ten traditional famous flowers in China. However, due to its short flowering period and small differences in flower type and color, it is difficult to identify different Osmanthus varieties through the traditional morphological strategy. In this study, SSR primers were developed based on the whole genome sequence of O. fragrans, which provides markers for variety identification and genetic diversity analysis of O. fragrans.【Method】Utilizing the published whole-genome data of O. fragrans, genome-wide SSR loci were systematically identified using MISA software. The distribution patterns, density, and motif characteristics (repeat unit length, frequency, and type) of SSR loci were comprehensively analyzed. Primer 3 software was then employed to design primers targeting polymorphic SSR regions. Subsequently, the polymorphism and stability of 131 pairs of primers were further verified. 【Result】A total of 180 838 SSR loci were obtained from the whole genome sequence of O. fragrans, with an average of one locus per 4.10 kb. Mononucleotide (42.59%), dinucleotide (39.03%), and trinucleotide (14.13%) repeats dominated the SSR types. Dinucleotide motifs were primarily AT (49.72%) and AG (46.68%), while trinucleotide repeats were predominantly composed of AAT (28.67%), ACC (18.86%), and AAG (13.37%). The 131 pairs of primers were sequenced by capillary electrophoresis in 32 varieties of four groups of Albus Group, Luteus Group, Aurantiacus Group and Asiaticus Group, and 16 pairs of primers were polymorphic; the number of alleles at a single locus was 4-16, and the average number of alleles at a single locus was 8.94.【Conclusion】The 16 newly developed SSR markers exhibit high polymorphism and reliability, providing an effective molecular toolkit for cultivar authentication, genetic diversity assessment, and fingerprint database establishment in O. fragrans.
| [1] |
臧德奎, 向其柏, 刘玉莲, 等. 中国桂花的研究历史、现状与桂花品种国际登录[J]. 植物资源与环境学报, 2003, 12(4):49-53.
|
| [2] |
向其柏, 刘玉莲. 桂花资源的开发与应用现状及发展趋势[J]. 南京林业大学学报(自然科学版), 2004, 28(增刊1):104-108.
|
| [3] |
臧德奎, 向其柏. 中国桂花品种分类研究[J]. 中国园林, 2004, 20(11):40-49.
|
| [4] |
|
| [5] |
丁刘慧子, 邳植, 吴则东. 甜菜品种SSR指纹图谱的构建及遗传多样性分析[J]. 作物杂志, 2021(5):72-78.
|
| [6] |
SOLMAZO,
|
| [7] |
张霞, 于卓, 金兴红, 等. 马铃薯SSR引物的开发、特征分析及在彩色马铃薯材料中的扩增研究[J]. 作物学报, 2022, 48(4):920-929.
|
| [8] |
|
| [9] |
|
| [10] |
段一凡, 王贤荣, 梁丽丽, 等. 桂花品种SSR荧光指纹图谱的构建[J]. 南京林业大学学报(自然科学版), 2014, 38(增刊1):1-6.
|
| [11] |
|
| [12] |
马寅峰. 桂花SSR引物的开发和SCoT分子标记体系的建立[D]. 开封: 河南大学, 2015.
|
| [13] |
李军, 董彬, 张超, 等. 桂花EST-SSR引物开发及在品种鉴定中的应用[J]. 浙江农林大学学报, 2018, 35(2):306-313.
|
| [14] |
|
| [15] |
孟清照, 李仕金, 董转年, 等. SSR引物开发方法概述[J]. 大众科技, 2007, 9(6):116-117,107.
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
原志敏. 玉米全基因组SSRs分子标记开发与特征分析[D]. 雅安: 四川农业大学, 2013.
|
| [29] |
孙陶泽, 母洪娜, 王良桂, 等. 桂花(Osmanthus fragrans)转录组SSR特征分析[J]. 分子植物育种, 2019, 17(7):2258-2263.
|
| [30] |
陈林, 李龙娜, 杨国栋, 等. 特有植物短丝木犀(Osmanthus serrulatus)转录组微卫星特征分析[J]. 分子植物育种, 2016, 14(4):959-965.
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
臧德奎, 向其柏. 桂花品种研究(英文)[J]. 南京林业大学学报(自然科学版), 2004,(S1):7-13.
|
/
| 〈 |
|
〉 |