 PDF(2336 KB)
PDF(2336 KB)


Genetic diversity and genetic structure analysis of principal Pseudotaxus chienii natural populations in Zhejiang Province
CAO Sen, GAO Kai, LIU Lingjuan, FANG Wanli, HE Xuekai, ZHOU Zhichun
Journal of Nanjing Forestry University (Natural Sciences Edition) ›› 2025, Vol. 49 ›› Issue (6) : 115-124.
 PDF(2336 KB)
PDF(2336 KB)
 PDF(2336 KB)
PDF(2336 KB)
Genetic diversity and genetic structure analysis of principal Pseudotaxus chienii natural populations in Zhejiang Province
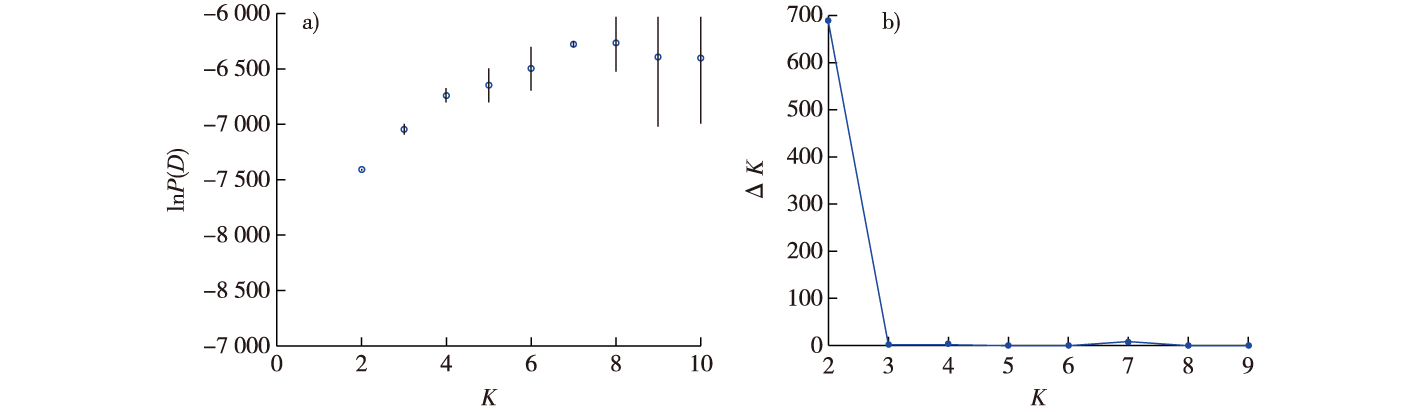
【Objective】The conservation of plant species with limited distribution and endangered status is a critical concern in biodiversity preservation. Among such species, Pseudotaxus chienii, a nationally protected plant species, has attracted significant attention due to its narrow habitat range and its vulnerability to extinction. As an endemic species primarily found in the mountainous regions of Zhejiang and Jiangxi provinces in China, P. chienii plays an important ecological role in maintaining forest biodiversity and stabilizing the local ecosystem. Unfortunately, over the past few decades, the natural populations of P. chienii have been severely affected by habitat destruction, fragmentation, and anthropogenic pressures, such as overharvesting and deforestation. These factors have contributed to a significant decline in both the population size and genetic diversity of this species. The research also sought to provide a deeper understanding of the evolutionary processes shaping the genetic makeup of P. chienii, especially in light of its restricted geographic range. By examining the genetic variation within and among populations, this study aimed to determine whether there are significant genetic differences that could have implications for long-term species survival. Additionally, the study aimed to investigate the extent of inbreeding and heterozygosity levels within these populations, which are important indicators of the overall fitness and adaptability of the species. The ultimate goal of the research was to generate critical genetic data that would serve as a foundation for future conservation and population restoration efforts, enabling the development of strategies that could enhance the species' genetic diversity and resilience in the wild.【Method】To achieve these objectives, a total of 133 individuals were collected from six natural populations of P. chienii located in different regions of Zhejiang Province (Fengyang Mountain Nature Reserve) and Quzhou City (Tianji Longmen Mountain Range). These populations were chosen for their geographic diversity, allowing for an examination of genetic variation across a range of environmental conditions and altitudes. In order to assess the genetic diversity and population structure of P. chienii, simple sequence repeat (SSR) molecular markers were employed. SSR markers are highly effective in detecting genetic variation and are widely used in plant population genetics studies. A total of 14 SSR primer pairs, known for their high polymorphism and ability to amplify variable loci, were selected for genotyping the 133 individuals. These markers were used to analyze a wide range of genetic parameters, including the number of alleles, the effective number of alleles (Ne), the Shannon's diversity index (I), and polymorphic information content (PIC). These parameters are commonly used to assess the level of genetic diversity within populations. Additionally, observed and expected heterozygosity (Ho and He) were calculated to evaluate the extent of genetic variation at the individual level, while fixation coefficients (F) and inbreeding coefficients (Fis) were determined to identify potential inbreeding and genetic drift within the populations. Furthermore, to explore the genetic structure of P. chienii, both principal coordinate analysis (PCoA) and unweighted pair group method with arithmetic mean (UPGMA) clustering were applied. These multivariate statistical methods allowed for the visualization of genetic relationships among individuals and provided insights into the clustering patterns of populations based on genetic similarities. By conducting these analyses, the study aimed to reveal any genetic subgroups within the species and identify the factors that contribute to the observed genetic structure. In summary, this study used advanced molecular techniques to investigate the genetic diversity and structure of P. chienii populations. By analyzing a large number of individuals from multiple natural populations, the study aimed to provide a comprehensive picture of the genetic variation within the species, and to understand the evolutionary and ecological factors that contribute to this variation.【Result】Significant differences were observed in tree height and diameter at breast height among the six populations of P. chienii, with the FYS2 population exhibiting the best growth status. Using 14 pairs of SSR primers with good polymorphism, genotyping of 133 P. chienii individuals identified a total of 102 alleles. The mean values of effective number of alleles (Ne), Shannon's diversity index (I), and polymorphic information content (PIC) were 4.69, 1.61 and 0.86, respectively. The average values of observed heterozygosity (Ho), expected heterozygosity (He), inbreeding coefficient (Fis) and fixation coefficient (F) were 0.34, 0.73, 0.54 and 0.58, respectively. Overall, the results indicated high genetic diversity in P. chienii populations, but there was a moderate level of heterozygote deficiency and obvious inbreeding within populations. Genetic structure analysis divided P. chienii individuals into two subgroups, which was also supported by PCoA and UPGMA clustering analysis.【Conclusion】The natural populations of P. chienii exhibit high levels of genetic diversity, with significant differences in genetic diversity among populations. Proximity-based clustering analysis reveals that geographically adjacent natural populations of P. chienii tend to cluster into the same subgroups, indicating that geographic variation is the primary factor influencing differences in the genetic structure in P. chienii.
Pseudotaxus chienii / nature populations / SSR markers / genetic diversity / genetic structure / Zhejiang Province
| [1] |
郑万钧, 傅立国. 中国植物志(第七卷)[M]. 北京: 科学出版社, 1978.
|
| [2] |
傅星, 南寅镐. 科尔沁沙地盐生草甸主要植物群落种群格局的研究[J]. 应用生态学报, 1992, 3(4):313-320.
The population patterns of main communities on halomorphic meadow of Keerqin sandyland and their formations are studied with methods of variance/mean, Greig-Smith's variance analysis for nested quadrats and Hill's pattern analysis. The results show that most of the species in <em>Aneurolepidium ehinense</em> and <em>Arundinella hirta communities</em> and all species in ecotone of these two communities are appeared as aggregated distribution. Its smallest scale is 0.01m<sup>2</sup>, and the biggest is 6.4×6.4m<sup>2</sup>. The minimum sampling size of grid should be less than 0.05×0.05m<sup>2</sup>, and the optimal sampling size is 40.96m<sup>2</sup>. The small scale pattern is caused by seed dispersion and nutrient reproduction, the middie scale one is caused by extension of stolon and long rhizome,and the large scale pattern is determined by total salt content of soil and soil pH.
|
| [3] |
徐晓婷, 杨永, 王利松. 白豆杉的地理分布及潜在分布区估计[J]. 植物生态学报, 2008, 32(5):1134-1145.
利用Diva-Gis软件对东亚特有单种属植物白豆杉(Pseudotaxus chienii)的地理分布格局进行研究。结合海拔图层和植被图层绘制白豆杉的分布图, 并用Diva-Gis中整合的Bioclim和Domain两个生态模型估测了白豆杉的潜在分布区。结果表明, 白豆杉分布在长江以南地区中低海拔450~1 500 m山区山坡林下、沟谷地带及溪边灌丛中, 浙江地区为白豆杉分布最为密集的地区。白豆杉的分布与植被类型和海拔有着密切的关系, 分布区的植被类型为常绿灌木林、阔叶林及常绿针阔叶混交林; 由东到西, 分布的最低海拔升高, 海拔范围缩小, 白豆杉资源量减少。白豆杉潜在分布区为浙、闽、粤、赣、湘、桂及黔几省及其交界处, 从浙江南部山区沿武夷山山系至南岭山系(向北到湘赣交界的罗宵山系甚至可以延伸到大别山系)至大瑶山山系(向北沿雪峰山)。保护方式应以就地保护为主, 同时兼顾其未来潜在的分布区, 关键是对白豆杉的适生区生境的保护。
|
| [4] |
胡绍庆, 陈征海, 孙孟军. 浙江省白豆杉资源调查研究[J]. 浙江大学学报(农业与生命科学版), 2003, 29(1):97-102.
|
| [5] |
王康, 杨永. 红豆杉科白豆杉属植物的分类学研究[J]. 植物分类学报, 2007, 45(6):862-869.
|
| [6] |
杨旭, 于明坚, 丁炳扬, 等. 凤阳山白豆杉种群结构及群落特性的研究[J]. 应用生态学报, 2005, 16(7):1189-1194.
对凤阳山自然保护区白豆杉(Pseudotaxus chienii)种群大小级结构、分布格局及群落特性进行了研究.结果表明,白豆杉种群在福建柏-猴头杜鹃林及猴头杜鹃矮林中生长最为良好,其重要值分别可达5%~10%.在猴头杜鹃矮林中,大小级为增长型,存活曲线为Deevy-Ⅰ型,种群将得到长期发展;在福建柏-猴头杜鹃林中,大小级为衰退型,存活曲线为Deevy-Ⅲ型,但由于幼苗储备量较为丰富,在一定时期内还将保持稳定,在这两种群落类型中,种群呈现集群分布.在常绿阔叶林中,白豆杉的重要值只有0.9%,并且种群大小级为衰退型,存活曲线为Deevy-Ⅱ型,分布格局为随机分布,说明常绿阔叶林不适合白豆杉种群的生长.通过对白豆杉和其他种群的相关分析表明,白豆杉种群与猴头杜鹃、福建柏等呈正相关,而与褐叶青冈、木荷等呈负相关.可以认为,如果凤阳山白豆杉种群的生境能够得到切实的保护,该种群就能长期存活和发展.
<i>Pseudotaxus chienii</i>,an endemic plant in China,is one of the second grade state protection wild plants,and distributes in Fengyangshan Natural Reserve as one of its concentrative dwelling places.A survey in the region was carried out on 10 communities,which were dominated by <i>P.chienii</i>.The analysis on its size structure,spatial distribution pattern and community characteristics showed that the populations of <i>P.chienii</i> could grow in the communities dominated by <i>Rhododendron simiarum</i>,Fokienia hodginsii-<i>R.simiarum</i>,and evergreen broad-leaved forests.In <i>R.simiarum</i> communities,the size structure,survival curve,and overwhelming community distribution pattern of <i>P.chienii</i> showed a sustaining development,while in communities dominated by <i>F.hodginsii</i>-<i>R.simiarum</i>,though the size structure was declining,the survival curve was Deevy-Ⅲ type.Plenty of plantlets were still existed,and the populations kept steady.Two types were considered to be the most suitable ones for <i>P.chienii</i> populations.The important value of <i>P.chienii</i> reached 5%~10%. In evergreen broad-leaved forests,the populations of <i>P.chienii</i> showed to be a declining type,and the survival curve was Deevy-Ⅱ type.The distribution pattern was random,indicating that <i>P.chienii</i> populations could not fit for survive in this kind of communities.Correlation analysis showed that <i>P.chienii</i> populations had a positive correlation with <i>R.simiarum</i> and <i>F.hodginsii</i>,but a negative correlation with <i>Cyclobalanopsis stewardiana</i> and <i>Schima superba</i>.<i>P.chienii</i> could associate to the habits with the community canopy density of about 0.6~0.8.From the results mentioned above,the populations of <i>P.chienii</i> could be able to sustain and develop,and the existing habitat in Fengyangshan should be protected effectively.The protection of <i>P.chienii</i> requires more basic work to establish efficient measures to protect its habitat.
|
| [7] |
杨旭. 凤阳山自然保护区白豆杉种群生态学的研究[D]. 杭州: 浙江大学, 2005.
|
| [8] |
李红英, 李康琴, 胥猛, 等. 北美鹅掌楸EST-SSR跨属间通用性[J]. 东北林业大学学报, 2011, 39(2):28-30,42.
|
| [9] |
黄秦军, 苏晓华, 张香华. SSR分子标记与林木遗传育种[J]. 世界林业研究, 2002, 15(3):14-21.
|
| [10] |
苏应娟, 王艇, 郑博, 等. 根据cpDNA trnL-F非编码区序列变异分析黑桫椤海南和广东种群的遗传结构与系统地理[J]. 生态学报, 2004, 24(5):914-919.
|
| [11] |
章艺, 刘鹏. 衢州珍稀濒危植物区系的研究[J]. 井冈山师范学院学报, 2002, 23(5):30-33.
|
| [12] |
李小白, 向林, 罗洁, 等. 转录组测序(RNA-seq)策略及其数据在分子标记开发上的应用[J]. 中国细胞生物学学报, 2013, 35(5):720-726,740.
|
| [13] |
王希, 陈丽, 赵春雷. 利用MISA工具对不同类型序列进行SSR标记位点挖掘的探讨[J]. 中国农学通报, 2016, 32(10):150-156.
为扩大MISA工具(MIcroSAtellite identification tool)的适用范围,本研究尝试用MISA对其他类型的序列进行分析,探讨关键参数对分析结果的影响。研究收集了5 种类型的甜菜序列数据,探讨了不同类型序列适用的相关分析参数,以及不同参数对分析结果的影响。利用MISA工具分析了5 种类型的甜菜序列,其中4 种来自公共数据库,1 种来自课题组前期工作。结果表明在适当的参数下,配合适当的序列库格式化处理,MISA可以应用于不同的甜菜序列类型。可分析的序列库长度上限取决于去冗余工具的要求,并影响去冗余处理的时间和结果,重复单元长度的限定参数影响位点挖掘数量和引物设计成功率,序列长度的分布范围不影响分析。应用参数调整后的脚本,从5 种甜菜序列库中共挖掘出了SSR标记位点约8000 个。可知MISA的应用范围可以扩展,需要调整的关键参数包括2 项:序列库的长度上限与重复单元长度。建议序列长度在程序与硬件允许的情况下尽量长,重复单元长度根据研究目标调整。
This work aims to extend the application range of MISA (MIcroSAtellite identification tool). The authors adjusted important parameters of MISA to find their effects on analysis results, and attempted to apply MISA to analyze other sequence types. Five different types of sequences of Beta vulgaris were collected and analyzed to find out appropriate parameters for each sequence type, and to clarify the effects of different parameters on analysis results. Among the five types of sequences, four were collected from public database and one was obtained from the early work of the research team. MISA was found to be able to analyze all these five sequence types of B. vulgaris sequences with appropriate analyzing parameters and sequence format. The max length of the sequence library was restricted by the redundant sequence removing tools, and affected redundant sequence processing, including both time consumption and analysis results. The length of detectable repeat units affected the amount of mined sites and the designing of primers. The length range of the sequence library did not disturb MISA analysis. About 8000 SSR sites were mined from the five types of B. vulgaris sequences using MISA tool with adjusted parameters. These results implied that MISA could be applied to analyzing different sequence types. Two parameters, the limit of sequence max length in the sequence pool and the length of detectable repeat units, should be adjusted. It is suggested that the sequences should be as long as possible under the permission of hardware and software, the length of repeat units should be adjusted according to the research purpose.
|
| [14] |
Summary: GenAlEx: Genetic Analysis in Excel is a cross-platform package for population genetic analyses that runs within Microsoft Excel. GenAlEx offers analysis of diploid codominant, haploid and binary genetic loci and DNA sequences. Both frequency-based (F-statistics, heterozygosity, HWE, population assignment, relatedness) and distance-based (AMOVA, PCoA, Mantel tests, multivariate spatial autocorrelation) analyses are provided. New features include calculation of new estimators of population structure: G′ST, G′′ST, Jost’s Dest and F′ST through AMOVA, Shannon Information analysis, linkage disequilibrium analysis for biallelic data and novel heterogeneity tests for spatial autocorrelation analysis. Export to more than 30 other data formats is provided. Teaching tutorials and expanded step-by-step output options are included. The comprehensive guide has been fully revised.
|
| [15] |
|
| [16] |
We describe a model-based clustering method for using multilocus genotype data to infer population structure and assign individuals to populations. We assume a model in which there are K populations (where K may be unknown), each of which is characterized by a set of allele frequencies at each locus. Individuals in the sample are assigned (probabilistically) to populations, or jointly to two or more populations if their genotypes indicate that they are admixed. Our model does not assume a particular mutation process, and it can be applied to most of the commonly used genetic markers, provided that they are not closely linked. Applications of our method include demonstrating the presence of population structure, assigning individuals to populations, studying hybrid zones, and identifying migrants and admixed individuals. We show that the method can produce highly accurate assignments using modest numbers of loci-e.g., seven microsatellite loci in an example using genotype data from an endangered bird species. The software used for this article is available from http://www.stats.ox.ac.uk/ approximately pritch/home. html.
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
阮咏梅, 张金菊, 姚小洪, 等. 黄梅秤锤树孤立居群的遗传多样性及其小尺度空间遗传结构[J]. 生物多样性, 2012, 20(4):460-469.
|
| [22] |
杨梅, 张敏, 师守国, 等. 武当木兰种群遗传结构的ISSR分析[J]. 林业科学, 2014, 50(1):76-81.
|
| [23] |
罗芊芊, 李峰卿, 肖德卿, 等. 两个南方红豆杉天然居群的交配系统分析[J]. 南京林业大学学报(自然科学版), 2023, 47(5):80-86.
【目的】研究南方红豆杉(Taxus wallichiana var. mairei)天然居群的交配系统,揭示浙江龙泉和江西分宜天然居群的交配机制以及亲代间的遗传多样性差异,为南方红豆杉的科学保育和遗传资源有效利用提供理论参考。【方法】以浙江龙泉和江西分宜天然居群亲代及其子代为研究对象,运用12对SSR引物对71个亲代及8个采种母株的104个子代进行分析。【结果】参试的浙江龙泉和江西分宜居群亲代的等位基因数(N<sub>a</sub>)皆为7,2个参试居群的有效等位基因数(N<sub>e</sub>)分别为4.831和3.552,浙江龙泉居群较江西分宜居群高出1.279个有效等位基因。2个参试居群的观测杂合度(H<sub>o</sub>)均小于期望杂合度(H<sub>e</sub>),且固定指数(F)大于0。浙江龙泉居群的等位基因丰富度和私有等位基因丰富度(A<sub>R</sub>=4.195,P<sub>A</sub>=1.832)皆大于江西分宜居群(A<sub>R</sub>=3.576,P<sub>A</sub>=1.647)。2个参试居群的遗传多样性略有差异,但仍具有较高的遗传多样性。多位点交配系统分析(MLTR)结果表明,浙江龙泉和江西分宜居群中,南方红豆杉的异交率较高(t<sub>m</sub>=1.200)。单位点和多位点父本相关性的差值[r<sub>p(s)</sub>-r<sub>p(m)</sub>]为0.095,亲代居群内存在近交现象,但近交水平较低( C t m - t s=0.268)。有效花粉供体(N<sub>ep</sub>=1.5)数目较少,表明南方红豆杉自然交配时亲本(父本)来源非常有限。【结论】南方红豆杉属于高度异交树种,2个参试居群的异交率差异不明显且遗传多样性皆较高,居群内存在近交现象,但近交水平较低。
|
| [24] |
程建慧, 廖荣俊, 宋其岩, 等. 基于SSR标记分析浙西南香椿天然居群的遗传多样性[J]. 中南林业科技大学学报, 2024, 44(2):146-156,165.
|
| [25] |
臧明月, 李璇, 方炎明. 基于SSR标记的白栎天然居群遗传多样性分析[J]. 南京林业大学学报(自然科学版), 2021, 45(1):63-69.
【目的】利用SSR分子标记,对白栎天然居群进行遗传多样性分析,为其种质资源的合理开发与保护提供指导。【方法】采用筛选出的SSR引物对3个群体进行遗传多样性与遗传结构研究,并计算遗传参数;基于个体间的遗传距离进行主成分分析,运用GenAlEx 6.5进行遗传距离与地理距离的相关性Mantel检验,软件Arlequin 用于分子方差分析,聚类分析采用MEGA 7中的UPGMA法;同时,使用Structure软件中的贝叶斯聚类算法探究群体遗传结构。【结果】6对SSR引物共检测到75个等位基因,平均有效等位基因数、观察杂合度和期望杂合度分别为7.16、0.70和 0.80,Shannon信息指数为2.05。AMOVA表明,群体内部的分子变异率高达97.41%,而群体之间的遗传变异仅为2.59%。黄山(YM)与天目山(TM)群体间的基因流为12.908,显著高于其他群体之间的交流水平,而二者间有着最近的遗传距离为0.272。贝叶斯法与UPGMA法聚类结果一致,表明来自YM与TM的个体亲缘关系更近。【结论】白栎具有较高的遗传多样性水平,且不同居群间的水平相当。YM和TM居群的基因交流最为密切,遗传分化不明显。白栎绝大部分遗传变异来源于群体内部,对于该物种的保护应减少不合理的人为开发行为,以原地保护为主,同时开展种质资源的收集与遗传改良工作。
|
| [26] |
胡绍庆. 浙江省白豆杉植物群落区系研究[J]. 浙江林业科技, 2001, 21(5):59-62.
|
| [27] |
林雪莹. 水松EST-SSR分子标记开发及群体遗传变异分析[D]. 长沙: 中南林业科技大学, 2018.
|
| [28] |
|
| [29] |
陈文文. 水杉(Metasequoia glyptostroboides)自然种群交配系统和扩散格局研究[D]. 上海: 华东师范大学, 2016.
|
| [30] |
邓莉丽, 刘青华, 周志春, 等. 抗松材线虫病马尾松种质资源遗传多样性分析及核心种质构建[J]. 浙江农林大学学报, 2024, 41(1):67-78.
|
| [31] |
魏利斌, 苗红梅, 李春, 等. 芝麻SNP和InDel标记遗传多样性、群体结构及连锁不平衡分析[J]. 分子植物育种, 2017, 15(8):3070-3079.
|
| [32] |
刘珊廷, 易显荣, 周民武, 等. 广西梨种质资源遗传多样性和群体结构分析[J]. 果树学报, 2024, 41(3):379-391.
|
| [33] |
杜康, 王加焕, 李俊恒, 等. 毛白杨耐寒种质资源遗传鉴定及评价[J]. 北京林业大学学报, 2023, 45(12):59-67.
|
| [34] |
罗芊芊. 南方红豆杉天然居群及家系遗传变异研究[D]. 长沙: 中南林业科技大学, 2020.
|
| [35] |
张俊红, 王洋, 周生财, 等. 闽楠群体遗传结构分析与核心种质库构建[J]. 林业科学, 2024, 60(1):68-79.
|
| [36] |
|
| [37] |
中国科学院华南研究所华南植物园, 广东省野生动植物保护管理办公室. 广东珍稀濒危植物[M]. 北京: 科学出版社, 2003.
South China Botanical Garden, South China Institute, Chinese Academy of Sciences,Guangdong Provincial Wildlife Conservation and Management Office. Rare and endangered plants in Guangdong[M]. Beijing: Science Press, 2003.
|
衢州市柯城区林业技术推广中心刘汝明和中国林业科学研究院亚热带林业研究所金国庆副研究员在材料收集与研究工作中给予了帮助。
/
| 〈 |
|
〉 |