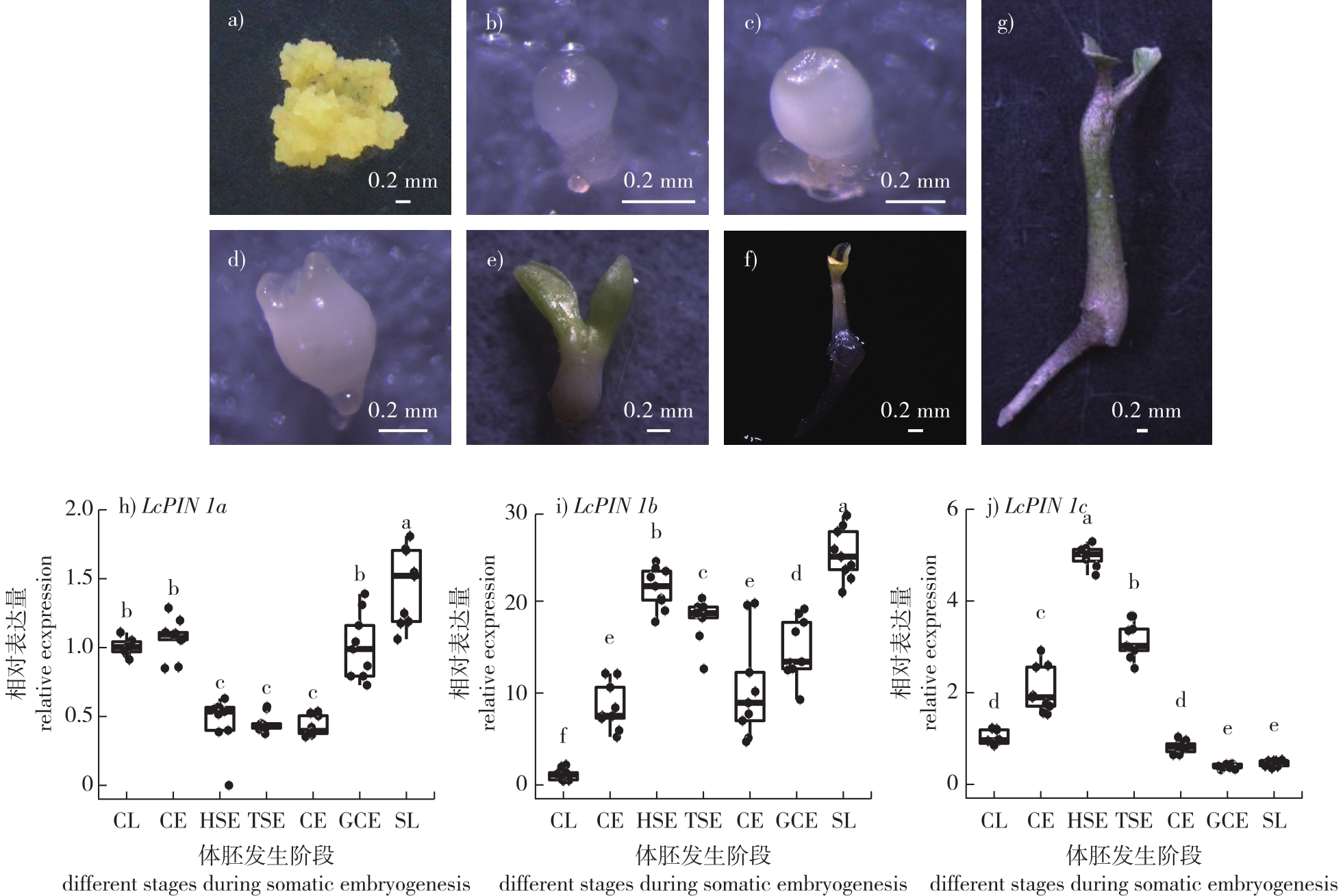
【Objective】This study aims to elucidate the expression patterns and functional roles of the auxin efflux carrier gene PIN1 during somatic embryogenesis in Liriodendron chinense.【Method】The temporal expression profiles of three homologous genes, LcPIN1a, LcPIN1b, and LcPIN1c, during somatic embryogenesis were analyzed using quantitative real-time PCR (qRT-PCR) at key developmental stages, including the globular, heart-shaped, torpedo-shaped, and cotyledon embryo stages. To investigate spatial expression, about 3.5 kb upstream promoter regions of each LcPIN1 gene were cloned and fused to the β-glucuronidase (GUS) reporter gene and the mCherry fluorescent protein reporter gene. The recombinant constructs were introduced into embryogenic callus of Liriodendron hybrid via Agrobacterium tumefaciens-mediated transformation. Transgenic calli were selected and induced to undergo somatic embryogenesis, followed by histochemical GUS staining and fluorescence microscopy to visualize the spatiotemporal expression patterns. In parallel, the full-length coding sequences of LcPIN1a, LcPIN1b and LcPIN1c were cloned into overexpression vectors driven by the constitutive Cauliflower Mosaic Virus 35S (CaMV35S) promoter and similarly transformed into embryogenic callus of Liriodendron hybrid. The effects of overexpression on somatic embryo formation were assessed by calculating the somatic embryogenesis efficiency, defined as the number of somatic embryos produced by a certain amount of embryogenic callus, and comparing the data statistically across independent transgenic lines.【Result】The qRT-PCR data revealed that all three LcPIN1 homologs were dynamically expressed during somatic embryogenesis, displaying both temporal overlap and distinct peaks at specific developmental stages. GUS staining and mCherry fluorescence assays provided high-resolution insights into their spatial expression patterns. During the globular embryo stage, LcPIN1a and LcPIN1c exhibited pronounced polar localization, with signals concentrated at one pole of the embryo, suggesting their early involvement in establishing embryonic polarity and the apical-basal axis. By contrast, LcPIN1b showed weak and diffuse expression at this stage. At the heart-shaped embryo stage, clear expression divergence was observed: LcPIN1a was predominantly expressed in the developing vascular tissues; LcPIN1b exhibited uniform expression throughout the embryo body; and LcPIN1c was specifically localized in the shoot apical meristem region and the nascent cotyledon primordia. As the embryos progressed into the torpedo-shaped and cotyledon stages, the three homologs displayed convergent expression patterns, with signals extending continuously from the cotyledon tips along the vascular strands down to the embryonic root tip. This suggests that LcPIN1a, LcPIN1b and LcPIN1c may coordinately maintain the auxin gradient required for proper somatic embryo elongation and differentiation during late embryogenesis. Overexpression experiments demonstrated that ectopic expression of each LcPIN1 homolog significantly inhibited somatic embryogenesis. Transgenic calli overexpressing LcPIN1a, LcPIN1b or LcPIN1c exhibited reduced somatic embryogenesis efficiency compared to wild-type controls, with a negative correlation between transgene expression levels and embryogenic potential. Notably, transgenic lines with higher levels of LcPIN1 overexpression showed a pronounced decline in somatic embryo formation, indicating that excessive auxin efflux may disrupt the finely tuned auxin gradients required for embryogenic competence and proper morphological development.【Conclusion】This study provides comprehensive evidence that the spatiotemporal precision of PIN1-mediated auxin transport plays a pivotal role in somatic embryogenesis of L. chinense. The redundant yet specialized expression patterns of LcPIN1a, LcPIN1b and LcPIN1c highlight their coordinated function in regulating auxin distribution at distinct developmental stages and tissue sites. Overexpression of LcPIN1 homologs perturbs auxin homeostasis, leading to impaired somatic embryogenesis. These findings advance our understanding of the molecular mechanisms underlying somatic embryogenesis in woody species and underscore the importance of auxin transport regulation for optimizing plant regeneration systems. This work also lays a foundation for future studies on functional diversification of PIN proteins and their potential applications in improving somatic embryogenesis efficiency in forest tree breeding.
 PDF(153890 KB)
PDF(153890 KB)


 PDF(153890 KB)
PDF(153890 KB)
 PDF(153890 KB)
PDF(153890 KB)