 PDF(17454 KB)
PDF(17454 KB)


Functional characterization of PjGRAS56, a transcription factor involved in shortened internode formation in Pseudosasa japonica var. tsutsumiana
LIU Shiying, JIANG Jiawen, GAO Zhipeng, WEI Qiang
Journal of Nanjing Forestry University (Natural Sciences Edition) ›› 2025, Vol. 49 ›› Issue (4) : 12-22.
 PDF(17454 KB)
PDF(17454 KB)
 PDF(17454 KB)
PDF(17454 KB)
Functional characterization of PjGRAS56, a transcription factor involved in shortened internode formation in Pseudosasa japonica var. tsutsumiana
【Objective】Internode length is a crucial trait for bamboo industrial utilization. Pseudosasa japonica var. tsutsumiana is a stable dwarf mutant of P. japonica, characterized by shortened, flask-shaped internodes, making it an ideal material for studying internode growth in bamboos. Preliminary studies identified the GRAS transcription factor PjGRAS56 as a key candidate gene responsible for the shortened internode formation in P. japonica var. tsutsumianais. This study aims to elucidate the potential role of PjGRAS56 in bamboo internode development.【Method】We employed bioinformatics to analyze the sequence features of PjGRAS56, confocal laser scanning microscopy to observe its subcellular localization, a yeast transcription system to assess its transcriptional activation activity, transgenic techniques in rice and Arabidopsis to analyze phenotypes of PjGRAS56-overexpressing plants, transcriptome sequencing and qRT-PCR to examine gene expression changes, and hand-sectioning to measure internode cell length.【Result】PjGRAS56 localized to the nucleus and cytoplasm, and lacked transcriptional activation activity. It was significantly down-regulated in the division zone of P. japonica var. tsutsumianais internodes. Overexpression of PjGRAS56 in rice resulted in dwarfism, shortened internodes, reduced tillering, shorter panicles and awns, fewer grains, and reduced leaf angle; overexpression of it in Arabidopsis led to smaller leaves and shorter flower stalks. Although the cell length in internodes of PjGRAS56-overexpressing plants was similar to controls, the cell number was significantly reduced. Transcriptome sequencing and differential gene expression analysis revealed significant down-regulation of cell division-related genes in PjGRAS56-overexpressing plants.【Conclusion】PjGRAS56 is a putative negative regulator of internode growth in bamboo.
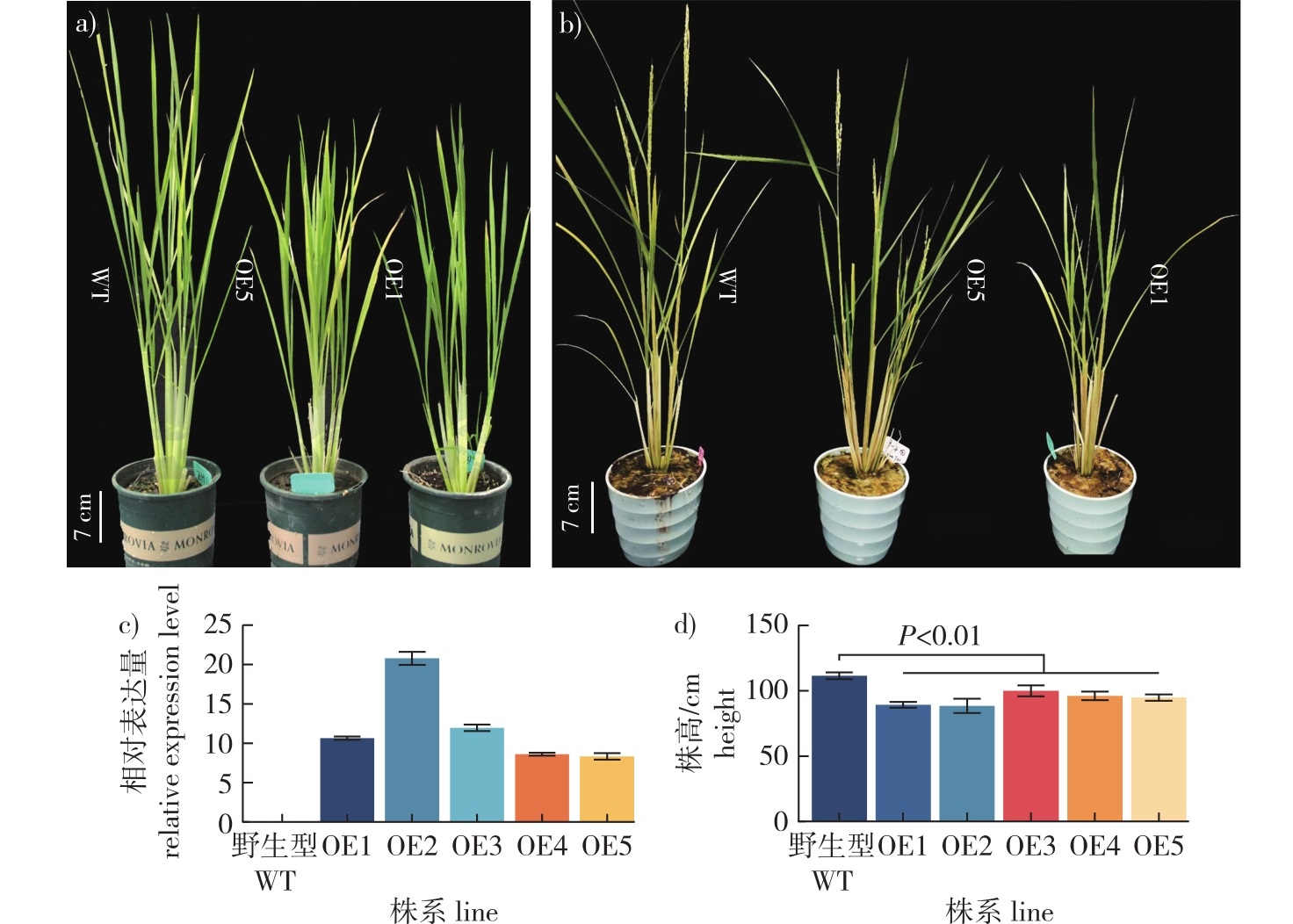
Pseudosasa japonica var. tsutsumiana / internode length / intercalary meristem / division zone / PjGRAS56
| [1] |
魏强, 郭琳, 陈铭, 等. 平安竹叶片缩小的细胞学观察[J]. 南京林业大学学报(自然科学版), 2019, 43(3):195-199.
|
| [2] |
林树燕, 丁雨龙. 平安竹抗旱生理指标的测定[J]. 林业科技开发, 2006(1):40-41.
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
吴占清, 陈威, 赵展, 等. 玉米GRAS基因家族的全基因组鉴定及生物信息学分析[J]. 中国农业科技导报, 2024, 26(3):15-25.DOI:10.13304/j.nykjdb.2023.0551.
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
却枫, 刘庆楠, 查若飞, 等. 孝顺竹中笋箨衰老相关WRKY转录因子的鉴定与分析[J]. 南京林业大学学报(自然科学版), 2023, 47(6):113-123.
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
童红宁. 水稻矮化少分蘖基因DLT的克隆鉴定及其在油菜素内酯信号传导中的功能分析[D]. 北京: 中国科学院大学, 2010.
|
| [26] |
苏强. GmFT1b/5b/3b基因调控大豆开花和生长发育的遗传效应分析[D]. 北京: 中国农业科学院, 2022.
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
杨建昌, 王朋, 刘立军, 等. 中籼水稻品种产量与株型演进特征研究[J]. 作物学报, 2006, 32(7):949-955.
|
| [35] |
彭显龙, 董强, 张辰, 等. 不同土壤条件下秸秆还田量对土壤还原性物质及水稻生长的影响[J]. 中国水稻科学, 2024, 38(2):198-210.
|
| [36] |
武志海, 徐克章, 赵颖君, 等. 吉林省47年来粳稻品种遗传改良过程中某些农艺性状的变化[J]. 中国水稻科学, 2007, 21(5):507-512.
|
| [37] |
高洪儒, 杨传铭, 赵北平, 等. 黑龙江省五常优质稻区2008—2022年水稻育种趋势分析[J]. 中国稻米, 2025, 31(3):87-94.
|
| [38] |
张耗, 谈桂露, 薛亚光, 等. 江苏省粳稻品种近60年演进过程中产量与形态生理特征的变化[J]. 作物学报, 2010, 36(1):133-140.
|
/
| 〈 |
|
〉 |