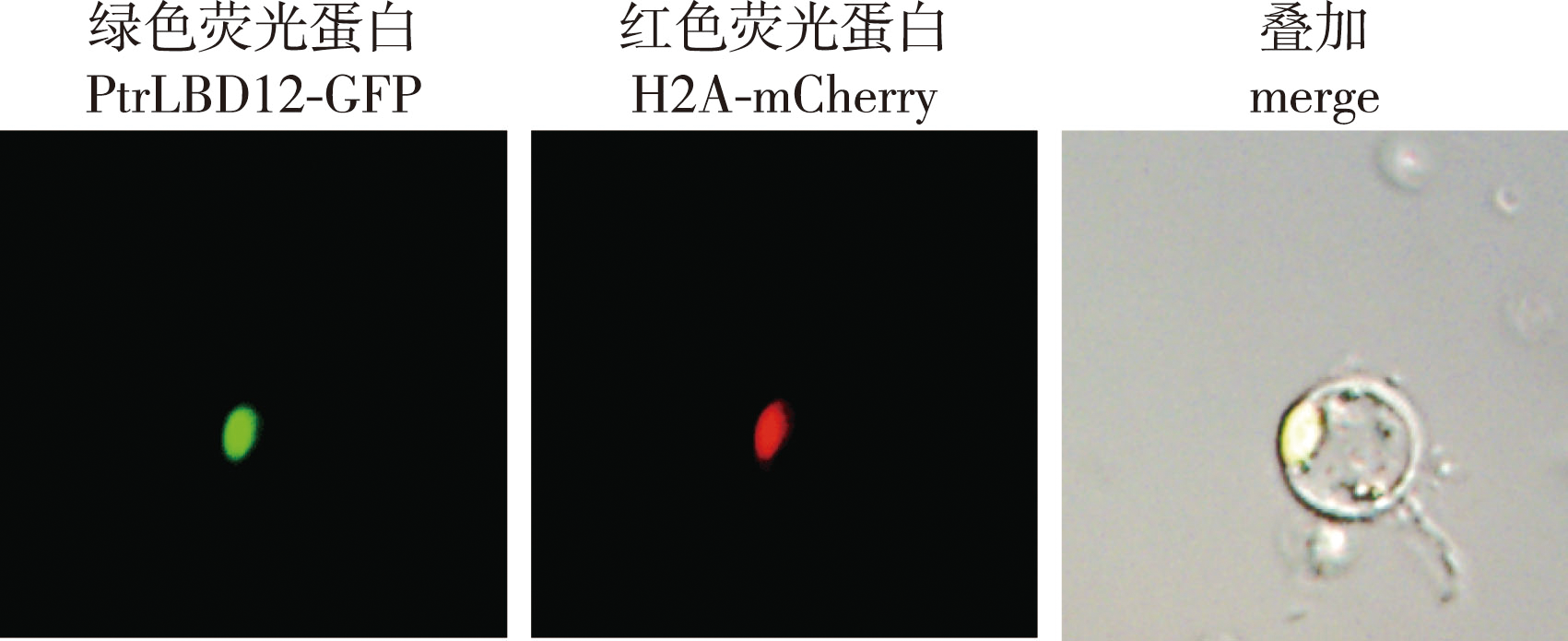
【Objective】The formation of wood, one of the most important raw materials for pulp and energy, depends on a complex and precise transcriptional regulation process in which transcription factors figure prominently. In Populus trichocarpa, PtrLBD12 (lateral organ boundaries domain 12) is a transcription factor lying downstream of PtrbHLH186, a key regulator of wood formation. Hence, PtrLBD12 was studied in depth here to investigate its function in tree growth and wood formation.【Method】 To determine the role of the PtrLBD12 transcription factor in the growth and wood formation of poplar trees, we analyzed its expression characteristics, generated PtrLBD12 overexpressing plants of P. trichocarpa, and assessed the growth and wood formation traits of transgenic plants. (1) Xylem, phloem, and terminal bud and leaf samples of wild-type P. trichocarpa plants, cultivated in a greenhouse, were collected to extract their respective RNA. Next, transcriptome sequencing was done to analyze the patterns of differentially expressed genes in those distinct tissues, as well as their expression levels of PtrLBD12. (2) To clarify the expression pattern of the PtrLBD12 protein, its subcellular localization was investigated by using the transient transformation system of stem-differentiating xylem protoplasts of P. trichocarpa. (3) To create PtrLBD12-overexpressing plants, the Agrobacterium tumefaciens-mediated transformation system of P. trichocarpa was used, with transgenic plants identified at both the DNA and RNA level. (4) Plant stem height, ground (basal) stem diameter, number of stem nodes, and length of the 8th stem node of transgenic and wild-type plants were measured at 30, 60 and 90 days after planting. (5) The 2nd, 4th, 6th and 8th stem segments of overexpressing and wild-type greenhouse plants cultivated for 4 months were paraffin-sectioned. Their stem characteristics were then respectively observed via Safranin O/Fast Green and Toluidine Blue staining. LAS X V2.0 software was used to calculate the number of vessel and fiber cells, as well as average lumen area of vessel. (6) The relative expression levels of 22 monolignol biosynthetic pathway genes in the PtrLBD12-overexpressing plants were determined by using quantitative PCR and applying the 2△△Ct calculation.【Result】 (1) Transcriptome analysis of the xylem, phloem, and terminal bud and leaf tissues of P. trichocarpa revealed a higher expression level of PtrLBD12 in both the xylem and phloem. (2) Subcellular localization showed that PtrLBD12 was expressed in the nucleus, where transcription factors in general were found. (3) Three PtrLBD12 overexpressing transgenic lines of P. trichocarpa were obtained, whose relative expression levels were 40.91, 79.51 and 102.19. (4) Overexpressing PtrLBD12 adversely affected the normal growth and development of P. trichocarpa: plant height, ground stem diameter, number of stem nodes, and length of the 8th stem node all decreased significantly in transgenic plants vis-à-vis the wild type. (5) However, overexpression of PtrLBD12 did lead to significantly more vessel and fiber cells per unit of area in the stems of transgenic plants, but these vessels had smaller lumen area. Furthermore, the augmented expression of PtrLBD12 enhanced lignification of the plant stem. (6) Finally, PtrLBD12’s overexpression bolstered the expression levels of multiple genes in the monolignol biosynthetic pathway, namely PtrPAL1, PtrC4H1, PtrC4H2, PtrHCT6, PtrCSE, PtrCSE2, PtrCCoAOMT1, PtrCCoAOMT2, PtrCCoAOMT3, PtrCCR2 and PtrCAld5H1.【Conclusion】As the gene downstream of PtrbHLH186, a key regulator of wood formation in poplar, the LBD12 transcription factor is able to govern the expression of monolignol biosynthetic genes, change the mode of lignin deposition, alter the morphology of xylem cells, and thereby affect plant growth and development.
 PDF(4417 KB)
PDF(4417 KB)


 PDF(4417 KB)
PDF(4417 KB)
 PDF(4417 KB)
PDF(4417 KB)