JOURNAL OF NANJING FORESTRY UNIVERSITY ›› 2024, Vol. 48 ›› Issue (2): 61-68.doi: 10.12302/j.issn.1000-2006.202302014
Previous Articles Next Articles
SONG Ziqi1,2( ), BIAN Guoliang1, LIN Feng3, HU Fengrong4, SHANG Xulan1,*(
), BIAN Guoliang1, LIN Feng3, HU Fengrong4, SHANG Xulan1,*( )
)
Received:2023-02-14
Revised:2023-03-23
Online:2024-03-30
Published:2024-04-08
CLC Number:
SONG Ziqi, BIAN Guoliang, LIN Feng, HU Fengrong, SHANG Xulan. Establishment and application of a flow cytometry method for chromosome ploidy identification of Cyclocarya paliurus[J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2024, 48(2): 61-68.
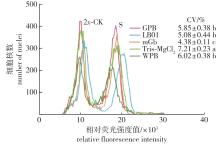
Fig. 1
Testing effects of ploidy analysis of Cyclocarya paliurus with different nuclei isolation buffers 2x-CK indicates the diploidy standard sample; S indicates the tetriploidy test sample; CV indicates the coefficient of variation in S; The different letters indicate that the significant differences among treatments (P<0.05). The same below."
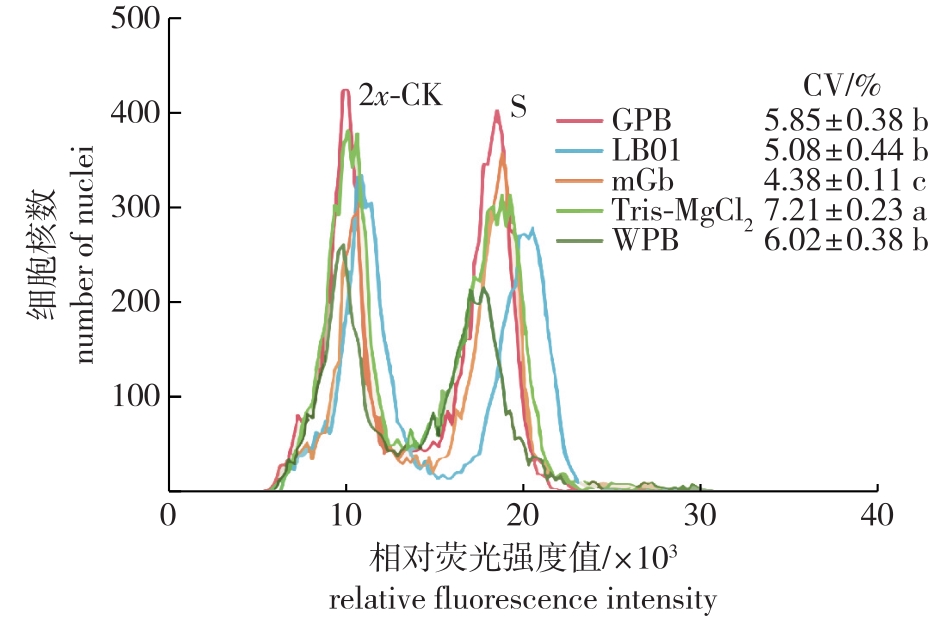
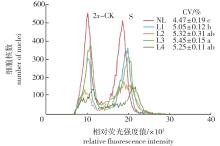
Fig. 2
Testing effect of ploidy analysis of C. paliurus with different centrifugal treatments L1 and L2 indicate that nuclei suspensions centrifuge at 1 000 r/min for 5 and 10 min, respectively; L3 and L4 indicate that nuclei suspensions centrifuge at 2 000 r/min for 5 and 10 min, respectively; NL indicates no centrifugal treatment."
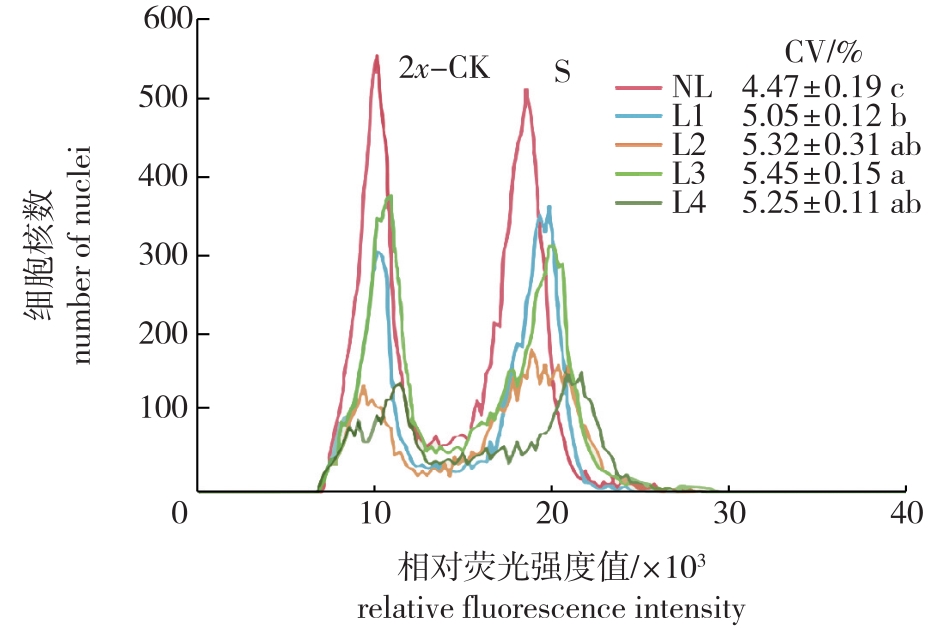
Table 2
Test results of ploidy of 1 395 C. paliurus germplasm resources by FCM"
| 来源 resource | 数量 number of samples | 倍性 ploidy | 占比/% proportion | 变异系数/% CV |
|---|---|---|---|---|
| 种质资源库 germplasm bank | 1 087 | 2 | 4.05 | 2.45~4.22 |
| 4 | 95.95 | 2.05~4.86 | ||
| 安徽省 Anhui Province | 75 | 2 | 38.67 | 3.02~5.54 |
| 4 | 61.33 | 2.76~5.21 | ||
| 湖北省 Hubei Province | 73 | 2 | 42.46 | 4.32~6.08 |
| 4 | 57.54 | 2.98~5.41 | ||
| 四川省 Sichuan Province | 32 | 4 | 100.00 | 3.42~5.03 |
| 江西省 Jiangxi Province | 80 | 4 | 100.00 | 3.10~5.41 |
| 广西壮族自治区 Guangxi Zhuang Autonomous Region | 48 | 4 | 100.00 | 2.68~4.97 |
| [1] | 方升佐. 青钱柳产业发展历程及资源培育研究进展[J]. 南京林业大学学报(自然科学版), 2022, 46(6): 115-126. |
| FANG S Z. A review on the development history and the resource silviculture of Cyclocarya paliurus industry[J]. J Nanjing For Univ (Nat Sci Ed), 2022, 46(6): 115-126. DOI: 10.12302/j.issn.1000-2006.202206019. | |
| [2] | 洪俊溪. 青钱柳人工林材性试验研究[J]. 福建林学院学报, 1997, 17(3): 214-217. |
| HONG J X. Experimental study on wood properties of Cyclocarya paliurus artificial forest[J]. J Fujian Coll For, 1997, 17(3): 214-217. | |
| [3] | ZHOU M M, LIN Y, FANG S Z, et al. Phytochemical content and antioxidant activity in aqueous extracts of Cyclocarya paliurus leaves collected from different populations[J]. Peer J, 2019, 7: e6492. DOI: 10.7717/peerj.6492. |
| [4] | YANG H M, YIN Z Q, ZHAO M G, et al. Pentacyclic triterpenoids from Cyclocarya paliurus and their antioxidant activities in FFA-induced HepG2 steatosis cells[J]. Phytochemistry, 2018, 151: 119-127. DOI: 10.1016/j.phytochem.2018.03.010. |
| [5] | FU X X, ZHOU X D, DENG B, et al. Seasonal and genotypic variation of water-soluble polysaccharide content in leaves of Cyclocarya paliurus[J]. South For, 2015, 77(3): 231-236. DOI: 10.2989/20702620.2015.1010698. |
| [6] | QU Y Q, SHANG X L, ZENG Z Y, et al. Whole-genome duplication reshaped adaptive evolution in a relict plant species, Cyclocarya paliurus[J]. Genomics Proteomics Bioinformatics, 2023:S1672-229(23)00033.DOI: 10.1016/j.gpb.2023.02.001. |
| [7] | SOLTIS P S, SOLTIS D E. The role of hybridization in plant speciation[J]. Annu Rev Plant Biol, 2009, 60(1): 561-588. DOI: 10.1146/annurev.arplant.043008.092039. |
| [8] | PIKAARD C S. Genomic change and gene silencing in polyploids[J]. Trends Genet, 2001, 17(12): 675-677. DOI: 10.1016/s0168-9525(01)02545-8. |
| [9] | 何法慧, 左倩倩, 于景金, 等. 35份狗牙根种质材料指纹图谱构建及染色体倍性鉴定[J]. 南京农业大学学报, 2023, 46(1):42-54. |
| HE F H, ZUO Q Q, YU J J, et al. Fingerprint construction and chromosome ploidy identification of 35 germplasms in bermudagrass[J]. J Nanjing Agric Univ, 2023, 46(1):42-54.DOI: 10.7685/jnau.202201035. | |
| [10] | 赵帅琪, 张伟伟, 牛俊芳, 等. 森林草莓和栽培草莓在果实发育和成熟过程中细胞壁变化的比较[J]. 植物生理学报, 2021, 57(12): 2323-2336. |
| ZHAO S Q, ZHANG W W, NIU J F, et al. Comparison of cell wall changes of Fragaria vesca and Fragaria × ananassa during fruit development and ripening[J]. Plant Physiol J, 2021, 57(12): 2323-2336. DOI: 10.13592/j.cnki.ppj.2021.0053. | |
| [11] | 康向阳. 杜仲良种选育研究现状及展望[J]. 北京林业大学学报, 2017, 39(3): 1-6. |
| KANG X Y. Status and prospect of improved variety selection in Eucommia ulmoides[J]. J Beijing For Univ, 2017, 39(3): 1-6. DOI: 10.13332/j.1000-1522.20160377. | |
| [12] | 李秀兰, 陈力. 三倍体丹参的培育及其可持续利用研究[J]. 中草药, 2012, 43(2): 375-379. |
| LI X L, CHEN L. Breeding for triploids of Salvia miltiorrhiza and its sustainable utilization[J]. Chin Tradit Herb Drugs, 2012, 43(2): 375-379. DOI: 10.7501/j.issn.0253-2670. | |
| [13] | DAS S K, SABHAPONDIT S, AHMED G, et al. Biochemical evaluation of triploid progenies of diploid×tetraploid breeding populations of Camellia for genotypes rich in catechin and caffeine[J]. Biochem Genet, 2013, 51: (5/6). DOI: 10.1007/s10528-013-9569-x. |
| [14] | XU C G, TANG T X, CHEN R, et al. A comparative study of bioactive secondary metabolite production in diploid and tetraploid Echinacea purpurea (L.) Moench[J]. Plant Cell Tiss Organ Cult, 2014, 116(3): 323-332. DOI: 10.1007/s11240-013-0406-z. |
| [15] | 陶抵辉, 李小红, 王利群, 等. 植物染色体倍性鉴定方法研究进展[J]. 生命科学研究, 2009, 13(5): 453-458. |
| TAO D H, LI X H, WANG L Q, et al. Progresses on determination of cell chromosome ploidy level of plants[J]. Life Sci Res, 2009, 13(5): 453-458. DOI: 10.16605/j.cnki.1007-7847.2009.05.011. | |
| [16] | SCHWARZACHER T, WANG, M L, LEITCH A R, et al. Flow cytometric analysis of the chromosomes and stability of a wheat cell-culture line[J]. Theor Appl Genet, 1997, 94(1): 91-97. DOI: 10.1007/s001220050386. |
| [17] | SLIWINSKA E. Flow cytometry:a modern method for exploring genome size and nuclear DNA synthesis in horticultural and medicinal plant species[J]. Folia Hortic, 2018, 30(1): 103-128. DOI: 10.2478/fhort-2018-0011. |
| [18] | 金亮, 徐伟韦, 李小白, 等. DNA流式细胞术在植物遗传及育种中的应用[J]. 中国细胞生物学学报, 2016, 38(2): 225-234. |
| JIN L, XU W W, LI X B, et al. Application of DNA flow cytometry to plant genetics and breeding[J]. Chin J Cell Biol, 2016, 38 (2): 225-234. DOI: 10.11844/cjcb.2016.02.0308. | |
| [19] | 宫雅昕, 岳涵, 向宇, 等. GABA代谢负调控叶片细胞内复制发生的机制研究[J]. 植物生理学报, 2020, 56(2): 235-246. |
| GONG Y X, YUE H, XIANG Y, et al. Mechanism study of negative regulation of GABA metabolism on endoreplication in Arabidopsis thaliana leaf development[J]. Plant Physiol J, 2020, 56(2): 235-246. DOI: 10.13592/j.cnki.ppj.2019.0452. | |
| [20] | GALBRAITH D W. Simultaneous flow cytometric quantification of plant nuclear DNA contents over the full range of described angiosperm 2C values[J]. Cytometry A, 2009, 75(8): 692-698. DOI: 10.1002/cyto.a.20760. |
| [21] | 田新民, 周香艳, 弓娜. 流式细胞术在植物学研究中的应用检测植物核DNA含量和倍性水平[J]. 中国农学通报, 2011, 27(9): 21-27. |
| TIAN X M, ZHOU X Y, GONG N, et al. Applications of flow cytometry in plant research-analysis of nuclear DNA content and ploidy level in plant cells[J]. Chin Agric Sci Bull, 2011, 27(9): 21-27. | |
| [22] | 韩杰, 沈海萍, 储冬生, 等. 4个薄壳山核桃品种核型分析[J]. 分子植物育种, 2018, 16(17): 5704-5711. |
| HAN J, SHEN H P, CHU D S, et al. Karyotype analysis of four pecan cultivars[J]. Mol Plant Breed, 2018, 16(17): 5704-5711. DOI: 10.13271/j.mpb.016.005704. | |
| [23] | 任伟超, 徐姣, 樊锐锋, 等. 应用流式细胞术对柳属染色体倍性与基因组大小测定[J]. 东北林业大学学报, 2021, 49(4): 56-61. |
| REN W C, XU J, FAN R F, et al. Chromosome ploidy and genome size determination of Salix using flow cytometry[J]. J Northeast For Univ, 2021, 49(4):56-61. DOI: 10.13759/j.cnki.dlxb.2021.04.010. | |
| [24] | PELLICER J, LEITCH I J. The application of flow cytometry for estimating genome size and ploidy level in plants[M]//Molecular Plant Taxonomy. Totowa, NJ: Humana Press, 2014: 279-307. DOI: 10.1007/978-1-62703-767-9_14. |
| [25] | DPOOLEŽEL J, BINAROVÁ P, LCRETTI S. Analysis of nuclear DNA content in plant cells by flow cytometry[J]. Biol Plant, 1989, 31(2): 113-120. DOI: 10.1007/BF02907241. |
| [26] | XU J, WEI X P, LIU J S, et al. Genome sizes of four important medicinal species in Kadsura by flow cytometry[J]. Chin Herb Med, 2021, 13(3): 416-420. DOI: 10.1016/j.chmed.2021.05.002. |
| [27] | SONG P, WANG X F, CAI M, et al. Research on identification of polyploids by flow cytometry in Lagerstroemia indica and Lagerstroemia subcostata[J]. Acta Hortic, 2012 (935): 207-212. DOI: 10.17660/actahortic.2012.935.29. |
| [28] | LOUREIRO J, RODRIGUEZ E, DOLEZEL J, et al. Flow cytometric and microscopic analysis of the effect of tannic acid on plant nuclei and estimation of DNA content[J]. Ann Bot-London, 2006, 98(3): 515-527. DOI: 10.1093/aob/mcl140. |
| [29] | 张桂芳, 王艳, 闫小巧, 等. 流式细胞仪检测铁皮石斛核DNA初探[J]. 现代中药研究与实践, 2017, 31(1): 16-19. |
| ZHANG G F, WANG Y, YAN X Q, et al. Study on flow cytometer for detecting nuclear DNA contents in Dendrobium officinal[J]. Res Pract Chin Med, 2017, 31(1): 16-19. DOI: 10.13728/j.1673-6427.2017.01.005. | |
| [30] | 于红梅, 王静, 赵密珍, 等. 利用流式细胞仪检测草莓倍性方法的优化[J]. 南方农业学报, 2012, 43(10): 1530-1533. |
| YU H M, WANG J, ZHAO M Z, et al. Optimization of strawberry ploidy identification method using flow cytometry[J]. J South Agric, 2012, 43(10): 1530-1533. DOI: 10.3969/j.issn.2095-1191.2012.10.1530. | |
| [31] | 杨静, 宋勤霞, 宁军权, 等. 利用流式细胞术鉴定桑树染色体倍性的方法[J]. 蚕业科学, 2017, 43(1): 8-17. |
| YANG J, SONG Q X, NING J Q, et al. Establishment of Morus L. chromosome ploidy identification method using flow cytometry[J]. Sci Seric, 2017, 43(1): 8-17. DOI: 10.13441/j.cnki.cykx.2017.01.002. | |
| [32] | 何婷, 郭桂梅, 陆瑞菊, 等. 两份大麦材料小孢子诱导愈伤及再生植株的倍性研究[J]. 植物生理学报, 2021, 57(8): 1708-1714. |
| HE T, GUO G M, LU R J, et al. Study on the ploidy of microspore-derived calli and regenerated plants of two barley materials[J]. Plant Physiol J, 2021, 57(8): 1708-1714. DOI: 10.13592/j.cnki.ppj.2021.0100. | |
| [33] | REUTEMANN A V, HONFI A I, KARUNARATHNE P, et al. Variation of residual sexuality rates along reproductive development in apomictic tetraploids of Paspalum[J]. Plants, 2022, 11(13): 1639. DOI: 10.3390/plants11131639. |
| [34] | PLASCHIL S, ABEL S, KLOCKE E. The variability of nuclear DNA content of different Pelargonium species estimated by flow cytometry[J]. PLoS One, 2022, 17(4): e0267496. DOI: 10.1371/journal.pone.0267496. |
| [35] | VIRUEL J, CONEJERO M, HIDALGO O, et al. A target capture-based method to estimate ploidy from herbarium specimens[J]. Front Plant Sci, 2019, 10: 937. DOI: 10.3389/fpls.2019.00937. |
| [36] | TOMASZEWSKA P, PELLNY T K, HERNÁNDEZ L M, et al. Flow cytometry-based determination of ploidy from dried leaf specimens in genomically complex collections of the tropical forage grass Urochloas L.[J]. Genes, 2021, 12(7): 957. DOI: 10.3390/genes12070957. |
| [37] | 李雯雯, 刘立强, 帕米尔·艾尼, 等. 利用流式细胞术鉴定新疆野杏染色体倍性和DNA含量[J]. 农业生物技术学报, 2019, 27(3): 542-550. |
| LI W W, LIU L Q, AINI P M E, et al. Identification of chromosomal ploidy and DNA content in Xinjiang Armeniaca vulgaris by flow cytometry[J]. J Agric Biotechnol, 2019, 27(3):542-550. DOI: 10.3969/j.issn.1674-7968.2019.03.019. | |
| [38] | CASTRO S, LOUREIRO J, RODRIGUEZ E, et al. Evaluation of polysomaty and estimation of genome size in Polygala vayredae and P. calcarea using flow cytometry[J]. Plant Sci, 2007, 172(6): 1131-1137. DOI: 10.1016/j.plantsci.2007.03.002. |
| [39] | GEORGIEV V, WEBER J, BLEY T, et al. Improved procedure for nucleus extraction for DNA measurements by flow cytometry of red beet (Beta vulgaris L.) hairy roots[J]. J Biosci Bioeng, 2009, 107(4): 439-441. DOI: 10.1016/j.jbiosc.2008.12.023. |
| [40] | 吕顺, 任毅, 王芳, 等. 利用流式细胞术快速鉴定169份香蕉种质资源的染色体倍性[J]. 果树学报, 2018, 35(6): 668-684. |
| LÜ S, REN Y, WANG F, et al. Ploidy identification of 169 Musa germplasms by flow cytometry[J]. J Fruit Sci, 2018, 35(6): 668-684. DOI: 10.13925/j.cnki.gsxb.20170419. |
| [1] | LIU Xiaofang, YUE Xiliang, FANG Shengzuo, LI Qing, SUN Xin. Effects of various ratios of nitrogen and phosphorus addition on the growth and contents of leaf bioactive substances in Cyclocarya paliurus [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2024, 48(4): 57-66. |
| [2] | LIU Li, QU Yinquan, YU Yanhao, WANG Qian, FU Xiangxiang. Analysis of SSR locus based on the whole genome sequences of Cyclocarya paliurus and the development of polymorphic primers [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2024, 48(4): 67-75. |
| [3] | LIU Xialan, SONG Ziqi, HU Fengrong, SHANG Xulan. A comparative study on leaf characters between diploid and tetraploid of Cyclocarya paliurus [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2024, 48(4): 76-84. |
| [4] | ZHANG Zanpei, GU Yueying, SHANG Xulan, WANG Ji, FANG Shengzuo. An evaluation on the cold tolerance of twenty-three Cyclocarya paliurus families under natural low temperatures [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2024, 48(4): 85-92. |
| [5] | WANG Ji, FANG Shengzuo. Effects of different anti-browning agents on enzyme activity and growth in callus of Cyclocarya paliurus [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2023, 47(6): 167-174. |
| [6] | HUANG Ziliang, XU Ziheng, SUN Caowen. Study on seasonal dynamics of seed rain and characteristics of soil seed banks in Cyclocarya paliurus [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2023, 47(2): 18-26. |
| [7] | MA Qiuyue, WANG Yuxiao, LI Qianzhong, LI Shushun, WEN Jing, ZHU Lu, YAN Kunyuan, DU Yiming, XIE Zhijun, LI Shuxian, OUYANG Fangqun, LU Chengdai. Estimation of genome sizes of six Acer species by flow cytometry and K-mer analysis [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2023, 47(1): 163-170. |
| [8] | FANG Shengzuo. A review on the development history and the resource silviculture of Cyclocarya paliurus industry [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2022, 46(6): 115-126. |
| [9] | XU Zhanhong, ZHU Ying, JIN Huiying, SUN Caowen, FANG Shengzuo. Variations in the contents of leaf pigments and polyphenols and photosynthesis traits in Cyclocarya paliurus with different leaf colors [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2022, 46(2): 103-110. |
| [10] | SUN Caowen, ZHONG Wenwen, FU Xiangxiang, SHANG Xulan, FANG Shengzuo. A study on growth and aboveground biomass production of juvenile Cyclocarya paliurus plantations [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2022, 46(1): 138-144. |
| [11] | DUAN Yifan, LI Lan, YANG Xinxin, WANG Xianrong, ZHANG Min, ZHANG Cheng, CHAI Zihan. Study on ploidy and genome sizes of Osmanthus fragrans and its related species [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2021, 45(5): 47-52. |
| [12] | LI Na, ZHU Peilin, FENG Cai, WEN Minxue, FANG Shengzuo, SHANG Xulan. Variations in physiological characteristics of rootstock-scion and its relationship to graft compatibility during the grafting union process of Cyclocarya paliurus [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2021, 45(1): 13-20. |
| [13] | ZHOU Yongsheng, XU Ziheng, YUAN Fayin, SHANG Xulan, SUN Caoweng, FANG Shengzuo. Comparisons of community characteristics among three natural forests of Cyclocarya paliurus in the subtropical region of China [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2021, 45(1): 29-35. |
| [14] | CHEN Pei, ZHOU Mingming, FANG Shengzuo, LIU Yang, YANG Wanxia, SHANG Xulan. Effects of light quality on accumulation of phenolic compounds and antioxidant activities in Cyclocarya paliurus (Batal.) Iljinskaja leaves from different families [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2020, 44(2): 17-25. |
| [15] | LIN Yuan, CHEN Pei, ZHOU Mingming, SHANG Xulan, FANG Shengzuo. Key bioactive substances and their antioxidant activities in Cyclocarya paliurus (Batal.) Iljinskaja leaves collected from natural populations [J]. JOURNAL OF NANJING FORESTRY UNIVERSITY, 2020, 44(2): 10-16. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||